User:Natronorubrum: Difference between revisions
No edit summary |
No edit summary |
||
| Line 2: | Line 2: | ||
Natronorubrum | Natronorubrum | ||
Cell structure, metabolism and life cycle | Cell structure, metabolism and life cycle | ||
=Ecology and pathogenesis= | =Ecology and pathogenesis= | ||
| Line 64: | Line 52: | ||
=Description and Significance= | =Description and Significance= | ||
Natronorubrum [Na.tro.no.rub’rum] was first proposed as a new genus in 1999 when traditional 16S rRNA sequence, and the additional DNA-DNA hybridization, lipid composition, and phylogenetic and phenotypic features identified it as a separate genus from other members of the family Halobacteriaceae[[#References|[11]]]. And five species were later identified within the genus Natronorubrum: N. bangense, N. tibetense, N. sulfidifaciens, N. aibiense, and N. sediminis. | Natronorubrum [Na.tro.no.rub’rum] was first proposed as a new genus in 1999 when traditional 16S rRNA sequence, and the additional DNA-DNA hybridization, lipid composition, and phylogenetic and phenotypic features identified it as a separate genus from other members of the family Halobacteriaceae[[#References|[11]]]. And five species [http://www.bacterio.cict.fr/n/natronorubrum.html] were later identified within the genus Natronorubrum: N. bangense, N. tibetense, N. sulfidifaciens, N. aibiense, and N. sediminis. | ||
Natronorubrum means “red of soda”, derived from the Arabic word “natrun” meaning “soda” or “sodium carbonate”, and the Latin word “rubrum” meaning “red”[[#References|[7]]] . The name accurately describes two key properties of Natronorubrum genus: salt-loving and red-purple morphology. Natronorubrum belongs to the class Halobacteria, which has only one order Halobacteriales and one family Halobacteriaceae[[#References|[11]]] , and it is comprised of the extremely halophilic members of the Archaea domain4. For example, this genus has a minimum salt requirement of 12% or 120g/L[[#References|[7]]] , which is equivalent to 4 times the salinity of seawater. In addition, Natronorubrum microbes form red-purple colonies of approximately 2mm in diameter[[#References|[11]]] . The bacterioruberin carotenoids present in the microorganisms is responsible for the red color3, and the color is a distinctive feature for many members of the family Halobacteriaceae. As a result, an abundance of halophilic Archaea present in hypersaline lakes often turns the water red[[#References|[9]]] (FIG 1). | Natronorubrum means “red of soda”, derived from the Arabic word “natrun” meaning “soda” or “sodium carbonate”, and the Latin word “rubrum” meaning “red”[[#References|[7]]]. The name accurately describes two key properties of Natronorubrum genus: salt-loving and red-purple morphology. Natronorubrum belongs to the class Halobacteria, which has only one order Halobacteriales and one family Halobacteriaceae[[#References|[11]]], and it is comprised of the extremely halophilic members of the Archaea domain4. For example, this genus has a minimum salt requirement of 12% or 120g/L[[#References|[7]]] , which is equivalent to 4 times the salinity of seawater. In addition, Natronorubrum microbes form red-purple colonies of approximately 2mm in diameter[[#References|[11]]]. The bacterioruberin carotenoids present in the microorganisms is responsible for the red color3, and the color is a distinctive feature for many members of the family Halobacteriaceae. As a result, an abundance of halophilic Archaea present in hypersaline lakes often turns the water red[[#References|[9]]] (FIG 1). | ||
The genus Natronorubrum is discovered in many hypersaline lakes. It could be a significant contributor to the hypersaline lake ecosystem as an aerobic chemoorganotroph, using oxygen as electron acceptor, and organic compounds as electron donor and source of energy. Natronorubrum Strain HG-1 of unclassified species can also oxidize thiosulfate (inorganic sulfur compounds) to tetrathionate during heterotrophic growth with acetate. This ability is widespread in Bacteria but previously found only in thermo-acidophilic Archaea. This reaction increases heterotrophic growth, by mechanisms not clearly understood, but thought to provide additional energy yields[[#References|[10]]] . The discovery of Natronorubrum contributes to our understanding of the Archaea domain and the alkaline lake ecosystem. | The genus Natronorubrum is discovered in many hypersaline lakes. It could be a significant contributor to the hypersaline lake ecosystem as an aerobic chemoorganotroph, using oxygen as electron acceptor, and organic compounds as electron donor and source of energy. Natronorubrum Strain HG-1 of unclassified species can also oxidize thiosulfate (inorganic sulfur compounds) to tetrathionate during heterotrophic growth with acetate. This ability is widespread in Bacteria but previously found only in thermo-acidophilic Archaea. This reaction increases heterotrophic growth, by mechanisms not clearly understood, but thought to provide additional energy yields[[#References|[10]]]. The discovery of Natronorubrum contributes to our understanding of the Archaea domain and the alkaline lake ecosystem. | ||
=Genome Structure= | |||
The complete genome for Natronorubrum has not been mapped at this current time. Only the DNA G+C content and the 16S rRNA partial sequences have been analyzed for each species. Natronorubrum is defined to have G+C content between 59.9-61.2mol%[[#References|[7]]]. N. sediminis, however, has G+C content of 61.9mol% and 62.5mol% for the two strains discovered[[#References|[5]]], exceeding the current description of Natronorubrum. Note that the publication on N. sediminis postdates the last amendment for the genus Natronorubrum. This demonstrates our limited knowledge of this genus and possible modifications to the genus description in the future. | |||
All 5 species’ 16S rRNA analyses have more than 95% consistency with each other[[#References|[5]]], which is the conventional method to map phylogenetic trees and within the acceptable value for genus classification. However, some scholars argue that based on 16s rRNA alone is not a completely accurate way to classify microorganisms. The DNA dependent RNA polymerase B’ subunit genomic sequencing might serve as a better phylogenetic map, because it is highly conserved and present in only one copy in Bacteria and Archaea, as opposed to multiple copies of 16s rRNA gene in some microorganisms. Natronorubrum tibetense might represent a new genus according to a recent study using this method because it shows less than 50% bootstrap support in the rpoB’ gene tree[[#References|[6]]]. Future full genome mapping might help to settle this debate. | |||
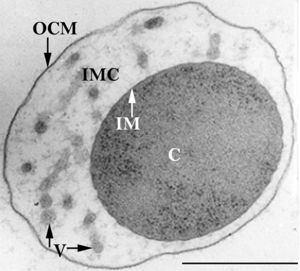
[[File:Huber-abb2.jpg|300px|thumb|left| Ultrathin section of an ''Ignicoccus hospitalis'' cell.]] | [[File:Huber-abb2.jpg|300px|thumb|left| Ultrathin section of an ''Ignicoccus hospitalis'' cell.]] | ||
= | =Cell Structure, Metabolism and Life Cycle= | ||
The | All Natronorubrum are pleomorphic, with triangular, square, polygonal or disc shapes, flat, and stains Gram-negative. The cells are 0.8-3.6um in size and form red colonies approximately 2mm in diameter after one week of growth in laboratory settings. Similar to many members of its family, Natronorubrum expresses C20-C25 and C20-C20 polar lipids, derivatives of phosphatidylglycerol (PG) and phosphatidylglycerol phosphate (PGP). They may also contain triglycerosyl diether-1 (TGD-1) and additional unidentified glycolipids in the membrane. Natronorubrum are strictly aerobic and chemoorganotrophic, and can be motile or non-motile. Natronorubrum can grow on a variety of sugars including glucose, sucrose and maltose, depending on the species. All are oxidase positive and catalase positive. Natronorubrum microbes are extreme halophilic, growing in at least 12% NaCl at pH 8-11. The cells lyse in distilled water. The cells grow at a temperature between 20°C and 55°C7, 11. Each Natronorubrum species has different optimal growth conditions within these ranges, as well as slightly different antibiotic resistance, cell structure and metabolism needs (Table 1). Unfortunately, no further information could be found at this time on the details of Natronorubrum life cycle, such as generation time, cell division, and proteins used for these processes. | ||
Strain information: Natronorubrum[http://www.straininfo.net/strains/search?taxon=Natronorubrum+sp.&includeSubtaxa=true&typeStrain=true] | |||
''Ignicoccus'' species are [http://en.wikipedia.org/wiki/Chemolithoautotroph chemolithoautotrophs] that use molecular hydrogen as the inorganic electron donor and elemental sulphur as the inorganic terminal electron acceptor[[#References|[1]]] . The reduction of the elemental sulphur results in the production of hydrogen sulphide gas. | ''Ignicoccus'' species are [http://en.wikipedia.org/wiki/Chemolithoautotroph chemolithoautotrophs] that use molecular hydrogen as the inorganic electron donor and elemental sulphur as the inorganic terminal electron acceptor[[#References|[1]]] . The reduction of the elemental sulphur results in the production of hydrogen sulphide gas. | ||
Revision as of 07:37, 14 December 2012
Natronorubrum
Cell structure, metabolism and life cycle
=Ecology and pathogenesis=
Natronorubrum strains are isolated from several saline lakes in Tibet, Xin-Jiang, Inner Mongolia of China and the Kulunda Steppe of Russia. Saline lakes are divided into aerobic and anaerobic layers. There are a dense population of cyanobacteria, and some aerobic organotrophs and aerobic sulfur oxidizers in the aerobic layer12. Natronorubrum, as a aerobic sulfur oxidizer and organotroph, is a contributing member of the sulfur and carbon cycle in the salt lake ecology. Microbes of the family, similar to Natronorubrum, are also found living in saline soils, subterranean salt deposits and solar salterns, and even saline microniches in non-saline environments3. This indicates the possibility of Natronorubrum bacteria being present in these environments as well.
One interesting environment that Natronorubrum is unable to survive on is Mars, as one research has proven. With the current interests towards finding microorganisms that can survive on Mars and the moon, some phylogenetically related halophilic microorganisms are reported to survive simulated Martian conditions. Natronorubrum’s ability to oxidize thiosulfate into tetrathionate would be advantageous to survival on sulfate heavy Mars. Unfortunately Natronorubrum strain HG-1 of unidentified species failed to survive under ultraviolet radiation, low temperatures, desiccation and exposure to Mars soil analogue8. This is to be expected since this genus is obligate aerobe.
Natronorubrum is not known to cause any disease within animals, plants, or humans.
=References= 1Cui, H-L., Tohty, D., Feng, J., Zhou, P-J., Liu, S-J. “Natronorubrum aibiense sp. nov. an extremely halophilic archaeon isolated from Aibi salt lake in Xin-Jiang, China, and emended description of the genus Natronobacterium.” Int J Syst Evol Microbiol, 2006, 56, 1515-1517. 2Cui, H-L., Tohty, D., Feng, Liu, H-C., Liu, S-J., Oren, A., J., Zhou, P-J. “Natronorubrum sulfudifaciens sp. nov. an extremely haloalkaliphilic archaeon isolated from Aiding salt lake in Xin-Jiang, China.” Int J Syst Evol Microbiol, 2007, 57, 738- 740. 3Fan, H.-P., Xue, Y.-F., Zeng, Y., Zhou, P.-J. & Ma, Y.-H. “Archaeal diversity of Zabuye lake in Tibet analyzed by culture- independent approach.” Wei Sheng Wu Xue Bao, 2003, 43, 401–408. 4Grant, W. D., Kamekura, M., McGenity, T. J. & Ventosa, A. “Order I. Halobacteriales Grant & Larsen 1989b, 495vp.” Bergey’s Manual of Systematic Bacteriology, 2001, 2 nd vol. 1, pp. 294-334. Edited by D. R. Boone, R. W. Castenholz & G. M. Garrity. New York: Springer. 5Gutiérrez, M. C., Castillo, A. M., Corral, P., Minegishi, H., Vantosa, A. “Natronorubrum sediminis sp. Nov., an archaeon isolated from a saline lake.” Int J Syst Evol Microbiol, 2009, 60, 1802-1806. 6Minegishi, H., Kamekura, M., Itoh, T., Echigo, A., Usami, R., Hashimoto, T. “Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B’ (rpoB’) gene.” Int J Syst Bacteriol, 2009, 60, 2398-2408. 7Oren, A., Arahal, D. R., Ventosa, A. “Emended descriptions of genera of the family Halobacteriaceae.” Int J Syst Evol Microbiol, 2009, 59, 637-642. 8Peeters, Z., Vos, D., ten Kate, I.L., Selch, F., van Sluis, C.A., Sorokin, D.Yu., Muijzer, G., Stan-Lotter, H., van Loosdrecht, M.C.M., Ehrenfreund, P. “Survival and death of the haloarchaeon Natronorubrum strain HG-1 in a simulated martian environment.” Adv. In Space Research, 2010, DOI: 10.1016/j.asr.2010.05.025 9Shahmohammadi, HR., Asgarani, E., Terato, H., Saito, T., Ohyama, Y., Gekko, K., Yamamoto, O., Ide, H. “Protective roles of bacterioruberin and intracellular KCl in the resistance of Halobacterium salinarium against DNA-damaging agents.” J Radiat Res, 1998, 39(4), 251-62. 10Sorokin, D. Y., Tourova, T. P. & Muyzer, G. “Oxidation of thiosulfate to tetrathionate by a haloarchaeon isolated from hyper- saline habitat.” Extremophiles, 2005, 9, 501–504. 11Xu, Y., Zhou, P. Tian, X. “Characterization of two novel haloalkaliphilic archaea, Natronorubrum bangense gen. nov., sp. nov. and Natronorubrum tibetense gen. nov., sp. nov.” Int J Syst Bacteriol, 1999, 49, 261-266. 12Jones, B., Grant, W., Duckworth, A., Owenson, G. “Microbial diversity of soda lakes.” Extremophiles, 1998, 2, 191-200.
Classification
Domain: Archaea
Kingdom: Euryarchaeota
Phylum: Euryarchaeota
Class: Halobacteria
Order: Halobacteriales
Family: Halobacteriaceae
Genus: Natronorubrum [1]
Species: N. aibiense, N. bangense, N. sulfidifaciens, N. tibetense, N. sediminis
NCBI taxonomy: Natronorubrum [2]
Description and Significance
Natronorubrum [Na.tro.no.rub’rum] was first proposed as a new genus in 1999 when traditional 16S rRNA sequence, and the additional DNA-DNA hybridization, lipid composition, and phylogenetic and phenotypic features identified it as a separate genus from other members of the family Halobacteriaceae[11]. And five species [3] were later identified within the genus Natronorubrum: N. bangense, N. tibetense, N. sulfidifaciens, N. aibiense, and N. sediminis.
Natronorubrum means “red of soda”, derived from the Arabic word “natrun” meaning “soda” or “sodium carbonate”, and the Latin word “rubrum” meaning “red”[7]. The name accurately describes two key properties of Natronorubrum genus: salt-loving and red-purple morphology. Natronorubrum belongs to the class Halobacteria, which has only one order Halobacteriales and one family Halobacteriaceae[11], and it is comprised of the extremely halophilic members of the Archaea domain4. For example, this genus has a minimum salt requirement of 12% or 120g/L[7] , which is equivalent to 4 times the salinity of seawater. In addition, Natronorubrum microbes form red-purple colonies of approximately 2mm in diameter[11]. The bacterioruberin carotenoids present in the microorganisms is responsible for the red color3, and the color is a distinctive feature for many members of the family Halobacteriaceae. As a result, an abundance of halophilic Archaea present in hypersaline lakes often turns the water red[9] (FIG 1).
The genus Natronorubrum is discovered in many hypersaline lakes. It could be a significant contributor to the hypersaline lake ecosystem as an aerobic chemoorganotroph, using oxygen as electron acceptor, and organic compounds as electron donor and source of energy. Natronorubrum Strain HG-1 of unclassified species can also oxidize thiosulfate (inorganic sulfur compounds) to tetrathionate during heterotrophic growth with acetate. This ability is widespread in Bacteria but previously found only in thermo-acidophilic Archaea. This reaction increases heterotrophic growth, by mechanisms not clearly understood, but thought to provide additional energy yields[10]. The discovery of Natronorubrum contributes to our understanding of the Archaea domain and the alkaline lake ecosystem.
Genome Structure
The complete genome for Natronorubrum has not been mapped at this current time. Only the DNA G+C content and the 16S rRNA partial sequences have been analyzed for each species. Natronorubrum is defined to have G+C content between 59.9-61.2mol%[7]. N. sediminis, however, has G+C content of 61.9mol% and 62.5mol% for the two strains discovered[5], exceeding the current description of Natronorubrum. Note that the publication on N. sediminis postdates the last amendment for the genus Natronorubrum. This demonstrates our limited knowledge of this genus and possible modifications to the genus description in the future.
All 5 species’ 16S rRNA analyses have more than 95% consistency with each other[5], which is the conventional method to map phylogenetic trees and within the acceptable value for genus classification. However, some scholars argue that based on 16s rRNA alone is not a completely accurate way to classify microorganisms. The DNA dependent RNA polymerase B’ subunit genomic sequencing might serve as a better phylogenetic map, because it is highly conserved and present in only one copy in Bacteria and Archaea, as opposed to multiple copies of 16s rRNA gene in some microorganisms. Natronorubrum tibetense might represent a new genus according to a recent study using this method because it shows less than 50% bootstrap support in the rpoB’ gene tree[6]. Future full genome mapping might help to settle this debate.
Cell Structure, Metabolism and Life Cycle
All Natronorubrum are pleomorphic, with triangular, square, polygonal or disc shapes, flat, and stains Gram-negative. The cells are 0.8-3.6um in size and form red colonies approximately 2mm in diameter after one week of growth in laboratory settings. Similar to many members of its family, Natronorubrum expresses C20-C25 and C20-C20 polar lipids, derivatives of phosphatidylglycerol (PG) and phosphatidylglycerol phosphate (PGP). They may also contain triglycerosyl diether-1 (TGD-1) and additional unidentified glycolipids in the membrane. Natronorubrum are strictly aerobic and chemoorganotrophic, and can be motile or non-motile. Natronorubrum can grow on a variety of sugars including glucose, sucrose and maltose, depending on the species. All are oxidase positive and catalase positive. Natronorubrum microbes are extreme halophilic, growing in at least 12% NaCl at pH 8-11. The cells lyse in distilled water. The cells grow at a temperature between 20°C and 55°C7, 11. Each Natronorubrum species has different optimal growth conditions within these ranges, as well as slightly different antibiotic resistance, cell structure and metabolism needs (Table 1). Unfortunately, no further information could be found at this time on the details of Natronorubrum life cycle, such as generation time, cell division, and proteins used for these processes.
Strain information: Natronorubrum[4]
Ignicoccus species are chemolithoautotrophs that use molecular hydrogen as the inorganic electron donor and elemental sulphur as the inorganic terminal electron acceptor[1] . The reduction of the elemental sulphur results in the production of hydrogen sulphide gas.
Ignicoccus are autotrophs in that they fix their own carbon dioxide into organic molecules. The carbon dioxide fixation process they use is a novel process called a dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle that involves 14 different enzymes[8] .
Members of the Ignicoccus genus are able to use ammonium as a nitrogen source.
Growth Conditions
Because members of the Ignicoccus genus are hyperthermophiles and obligate anaerobes, it is not surprising that their growth conditions are very complex. They are grown in a liquid medium known as ½ SME Ignicoccus which is a solution of synthetic sea water which is then made anaerobic.
Grown in this media at their optimal growth temperature of 90C, the members of the Ignicoccus genus typically reach a cell density of ~4x107cells/mL[1] .
The addition of yeast extract to the ½ SME media has been shown to stimulate the growth and increase maximum cell density achieved. The mechanism by which this is achieved is not known[1] .
Symbiosis
Ignicoccus hospitalis is the only member of the genus Ignicoccus that has been shown to have an extensive symbiotic relationship with another organism.
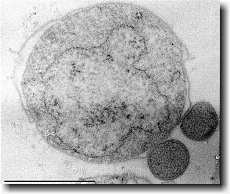
Ignicoccus hospitalis has been shown to engage in symbiosis with Nanoarchaeum equitans . Nanoarchaeum equitans is a very small coccoid species with a cell diameter of 0.4 µm[9] . Genome analysis has provided much of the known information about this species.
To further complicate the symbiotic relationship between both species, it’s been observed that the presence of Nanoarchaeum equitans on the surface of Ignicoccus hospitalis somehow inhibits the cell replication of Ignicoccus hospitalis . How or why this occurs has not yet been elucidated[3] .
Nanoarchaeum equitans
Nanoarchaeum equitans has the smallest non-viral genome ever sequenced at 491kb[9] . Analysis of the genome sequence indicates that 95% of the predicted proteins and stable RNA molecules are somehow involved in repair and replication of the cell and its genome[3] .
Analysis of the genome also showed that Nanoarchaeum equitans lacks nearly all genes known to be required in amino acid, nucleotide, cofactor and lipid metabolism. This is partially supported by the evidence that Nanoarchaeum equitans has been shown to derive its cell membrane from its host Ignicoccus hospitalis cell membrane. The direct contact observed between Nanoarchaeum equitans and Ignicoccus hospitalis is hypothesized to form a pore between the two organisms in order to exchange metabolites or substrates (likely from Ignicoccus hospitalis towards Nanoarchaeum equitans due to the parasitic relationship). The exchange of periplasmic vesicles is not thought to be involved in metabolite or substrate exchange despite the presence of vesicles in the periplasm of Ignicoccus hospitalis .
These analyses of the Nanoarchaeum equitans genome support the fact of the extensive symbiotic relationship between Nanoarchaeum equitans and Ignicoccus hospitalis. However, it has not yet been proven that it is a strictly parasitic relationship and further research may prove that there is a commensal relationship between the two species.
References
(1) Burggraf S., Huber H., Mayer T., Rachel R., Stetter K.O. and Wyschkony I. ” Ignicoccus gen. nov., a novel genus of hyperthermophilic, chemolithoautotrophic Archaea, represented by two new species, Ignicoccus islandicus sp. nov. and Ignicoccus pacificus sp. nov.” International Journal of Systematic and Evolutionary Microbiology, 2000, Volume 50.
(2) Naether D.J. and Rachel R. “The outer membrane of the hyperthermophilic archaeon Ignicoccus: dynamics, ultrastructure and composition.” Biochemical Society Transactions, 2004, Volume 32, part 2.
(3) Giannone R.J., Heimerl T., Hettich R.L., Huber H., Karpinets T., Keller M., Kueper U., Podar M. and Rachel R. “Proteomic Characterization of Cellular and Molecular Processes that Enable the Nanoarchaeum equitans- Ignicoccus hospitalis Relationship.” PLoS ONE, 2011, Volume 6, Issue 8.
(4) Eisenreich W., Gallenberger M., Huber H., Jahn U., Junglas B., Paper W., Rachel R. and Stetter K.O. “Nanoarchaeum equitans and Ignicoccus hospitalis: New Insights into a Unique, Intimate Association of Two Archaea.” Journal of Bacteriology, 2008, DOI: 10.1128/JB.01731-07.
(5) Grosjean E., Huber H., Jahn U., Sturt H, and Summons R. “Composition of the lipids of Nanoarchaeum equitans and their origin from its host Ignicoccus sp. strain KIN4/I.” Arch Microbiol, 2004, DOI: 10.1007/s00203-004-0725-x.
(6) Briegel A., Burghardt T., Huber H., Junglas B., Rachel R., Walther P. and Wirth R. “Ignicoccus hospitalis and Nanoarchaeum equitans: ultrastructure, cell–cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography.” Arch Microbiol, 2008, DOI 10.1007/s00203-008-0402-6.
(7) Burghardt T., Huber H., Junglas B., Naether D.J. and Rachel R. “The dominating outer membrane protein of the hyperthermophilic Archaeum Ignicoccus hospitalis: a novel pore-forming complex.” Molecular Microbiology, 2007, Volume 63.
(8) Berg I.A., Eisenreich W., Eylert E., Fuchs G., Gallenberger M., Huber H.,Jahn U. and Kockelkorn D. “A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis.” PNAS, 2008, Volume 105, issue 22.
(9) Brochier C., Gribaldo S., Zivanovic Y., Confalonieri F. and Forterre P. “Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales?” Genome Biology 2005, DOI:10.1186/gb-2005-6-5-r42.
(10) Huber H., Rachel R., Riehl S. and Wyschkony I. “The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon.” Archaea, 2002, Volume 1.