Antibiotics and the Human Gut Microbiota
Introduction
Antibiotics make up a class of molecules whose purpose is to inhibit the growth of or kill harmful bacteria. Antibiotics originate from a wide variety of microbes as a means of competing with and defending themselves from nearby bacteria and are used to treat bacterial infections in humans (1,2)[1][2]. A microbiome is a population that is defined by every microbe present on and within an organism; for humans, it contributes immensely to the physiological processes of the host organism (3)[3]. Although antibiotics are incredibly useful in fighting bacterial infections, they often alter the human gut microbiome in ways that prove detrimental to our health.
Common Uses for Antibiotics
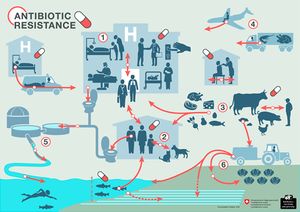
While antibiotics are primarily utilized to treat bacterial infections in humans, there are many other applications that significantly contribute to human antibiotic overexposure. Antibiotics are often used to treat and prevent bacterial infections in pets, livestock, and aquaculture (2)[4]. Additionally, antibiotics are used frequently to promote growth in livestock; certain studies estimate that the US uses more antibiotics for this purpose than for treatment of human bacterial infections (2c)[5]. Antibiotics are also included in many household products such as soaps, shampoos, and even solid objects such as fabrics (2)[4]. Aside from direct exposure, antibiotics and antibiotic resistant bacteria can come into contact with the human microbiota through waterway contamination (2a) and the consumption of food that is grown or produced in the presence of antibiotics (2b)[6].
Effect of Antibiotic Exposure on the Human Microbiota
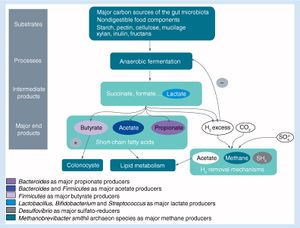
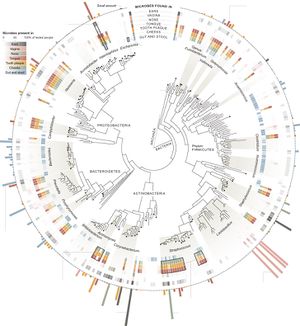
Due to the prevalence of antibiotic use throughout the world, the human microbiome is often exposed to excessive amounts of these antibacterial agents (2)[7]. Although antibiotics very effectively assist in the removal of harmful bacteria, they also have a significant effect on the gut microbiome. The most immediate of these effects is seen in the stress placed on the gut microbiome’s inhabitants, increasing expression of genes related to stress response, antibiotic resistance, and phage induction (2a)[8]. Bacterial fitness is altered in the presence of this stress, a change that tends to decrease bacterial diversity, richness and evenness within the gut; this disturbance is known as dysbiosis (2)[7]. Although the microbiome is generally able to return to a composition that is similar to that of pretreatment conditions (one study found this to occur about four weeks after five days of ciprofloxacin, although some taxa failed to recover until 6 months after treatment (5a)[9]), some alterations to the microbiome have been shown to persist over a number of months (2b,2c)[10][11]. It was also found that when exposed to targeted antibiotics, certain subpopulations existing outside of their range of effectiveness can still experience a reduction in population size (6)[12].
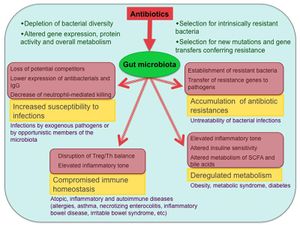
Antibiotic treatment and the resulting dysbiosis has been associated with a number of detrimental consequences. Although it is well known that antibiotic treatment leads to the selection for antibiotic resistance in pathogenic bacteria, this same type of selection occurs within the native inhabitants of the microbiome. Relatively high levels of antibiotic resistance genes have been shown to persist within the human gut microbiome for up to four years after antibiotic withdrawal (4)[13]. This is especially concerning when considering that such genes have been found to experience lateral transfer between distantly related bacterial species (2d)[14], allowing the gut microbiome to act as a kind of storehouse for antibiotic resistance genes that can theoretically be accessed by dangerous bacteria during future infections (2)[7]. A more immediate concern following antibiotic treatment is an increased vulnerability to infection. Both the reduction in diversity of the gut microbiome and its domination by antibiotic resistant strains of certain bacteria have been linked to dangerous post-treatment infections. For example, antibiotic treated mice have shown increased susceptibility to Clostridium difficile-induced Colitis (2f)[15] and bloodstream infection by vancomycin-resistant Enterococcus (2e)[16]. Early life dysbiosis resulting from antibiotic exposure has also been associated with an impaired immune response- leading to the development or enhancement of allergies, childhood asthma (2g)[17], and various inflammatory disorders such as late onset sepsis (2h)[18] and irritable bowel syndrome (2i)[19]. Treatment with antibiotics has been associated with increased weight gain in humans (2j)[20] and an increased risk of obesity in mice (2k)[21].
Avoiding Adverse Side Effects
There are many proposed methods of addressing these issues without full withdrawal from antibiotic use. Within the context of human treatment, probiotics show promise in being able to stabilize the microbiome and supplement the deficits produced by dysbiosis, such as in the case of streptomycin treated mice (5a)[22]. Additionally, the intake of certain ligands can signal bacterial gene expression that inhibits growth of other bacteria that tend to dominate the microbiome during and after treatment (5b)[23]. Synthetic medicinal compounds also show promise in being able to reduce the side effects of antibiotics without reducing their effectiveness, such as the DAV132 product created by Jean de Gunzberg and his research team to remove free moxifloxacin from the gut (2)[24]. Otherwise, limiting antibiotic use wherever possible will significantly reduce human contact and thus stifle the side effects of overexposure (3)[25].
Conclusion
The human gut microbiome plays an incredibly important role in the proper functioning of many physiological processes, most notably the human metabolism and immune system. Thus, the alteration of its composition that often accompanies exposure to antibiotics can aid in the development of various ailments, such as obesity and allergies, and reduce the effectiveness of our immune system. Antibiotic exposure along with subsequent selection for antibiotic resistance in pathogenic bacteria leads to the development and prevalence of fairly stable antibiotic resistance genes in native bacterial populations; this could allow future invaders to gain resistance through horizontal gene transfer. The wide range of uses for antibiotics means that humans are often overexposed to them. Treatment options such as probiotics can work to counteract the negative effects of antibiotic exposure, but the only available action that addresses all issues regarding overexposure is the more efficient and conservative use of antibiotics.
References
- ↑ “Microbes and the Human Body: Antibiotics.” Microbes and the Human Body | Microbiology Society, Microbiology Society, https://microbiologysociety.org/why-microbiology-matters/what-is-microbiology/microbes-and-the-human-body/antibiotics.html#:~:text=Antibiotics%20are%20chemicals%20that%20kill,by%20soil%20bacteria%20and%20fungi.
- ↑ Chandra N., Kumar S. (2017) Antibiotics Producing Soil Microorganisms. In: Hashmi M., Strezov V., Varma A. (eds) Antibiotics and Antibiotics Resistance Genes in Soils. Soil Biology, vol 51. Springer, Cham. https://doi.org/10.1007/978-3-319-66260-2_1
- ↑ Shreiner, A. B., Kao, J. Y., & Young, V. B. (2015). The gut microbiome in health and in disease. Current opinion in gastroenterology, 31(1), 69–75. https://doi.org/10.1097/MOG.0000000000000139
- ↑ 4.0 4.1 Meek, R. W., Vyas, H., & Piddock, L. J. (2015). Nonmedical Uses of Antibiotics: Time to Restrict Their Use?. PLoS biology, 13(10), e1002266. https://doi.org/10.1371/journal.pbio.1002266
- ↑ Landers, T. F., Cohen, B., Wittum, T. E., & Larson, E. L. (2012). A review of antibiotic use in food animals: perspective, policy, and potential. Public health reports (Washington, D.C. : 1974), 127(1), 4–22. https://doi.org/10.1177/003335491212700103
- ↑ Smith, K. E., Besser, J. M., Hedberg, C. W., Leano, F. T., Bender, J. B., Wicklund, J. H., ... & Osterholm, M. T. (1999). Quinolone-resistant Campylobacter jejuni infections in Minnesota, 1992–1998. New England Journal of Medicine, 340(20), 1525-1532.
- ↑ 7.0 7.1 7.2 Francino, M. P. (2016). Antibiotics and the human gut microbiome: dysbioses and accumulation of resistances. Frontiers in microbiology, 6, 1543.
- ↑ Maurice, C. F., Haiser, H. J., & Turnbaugh, P. J. (2013). Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell, 152(1-2), 39-50.
- ↑ Dethlefsen, L., Huse, S., Sogin, M. L., & Relman, D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS biol, 6(11), e280.
- ↑ De La Cochetière, M. F., Durand, T., Lepage, P., Bourreille, A., Galmiche, J. P., & Dore, J. (2005). Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. Journal of clinical microbiology, 43(11), 5588-5592.
- ↑ Jernberg, C., Löfmark, S., Edlund, C., & Jansson, J. K. (2007). Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. The ISME journal, 1(1), 56-66.
- ↑ Sekirov, I., Tam, N. M., Jogova, M., Robertson, M. L., Li, Y., Lupp, C., & Finlay, B. B. (2008). Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infection and immunity, 76(10), 4726-4736.
- ↑ Jakobsson, H. E., Jernberg, C., Andersson, A. F., Sjölund-Karlsson, M., Jansson, J. K., & Engstrand, L. (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PloS one, 5(3), e9836.
- ↑ De Vries, L. E., Valles, Y., Agersø, Y., Vaishampayan, P. A., Garcia-Montaner, A., Kuehl, J. V., ... & Francino, M. P. (2011). The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PloS one, 6(6), e21644.
- ↑ Buffie, C. G., Jarchum, I., Equinda, M., Lipuma, L., Gobourne, A., Viale, A., ... & Pamer, E. G. (2012). Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infection and immunity, 80(1), 62-73.
- ↑ Ubeda, C., Taur, Y., Jenq, R. R., Equinda, M. J., Son, T., Samstein, M., ... & Pamer, E. G. (2010). Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. The Journal of clinical investigation, 120(12), 4332-4341.
- ↑ McKeever, T. M., Lewis, S. A., Smith, C., Collins, J., Heatlie, H., Frischer, M., & Hubbard, R. (2002). Early exposure to infections and antibiotics and the incidence of allergic disease: a birth cohort study with the West Midlands General Practice Research Database. Journal of Allergy and Clinical Immunology, 109(1), 43-50.
- ↑ Mai, V., Torrazza, R. M., Ukhanova, M., Wang, X., Sun, Y., Li, N., ... & Neu, J. (2013). Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PloS one, 8(1), e52876.
- ↑ Durbán, A., Abellán, J. J., Jiménez‐Hernández, N., Salgado, P., Ponce, M., Ponce, J., ... & Moya, A. (2012). Structural alterations of faecal and mucosa‐associated bacterial communities in irritable bowel syndrome. Environmental Microbiology Reports, 4(2), 242-247.
- ↑ Angelakis, E., Armougom, F., Million, M., & Raoult, D. (2012). The relationship between gut microbiota and weight gain in humans. Future microbiology, 7(1), 91-109.
- ↑ Cho, I., Yamanishi, S., Cox, L., Methé, B. A., Zavadil, J., Li, K., ... & Li, H. (2012). Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature, 488(7413), 621-626.
- ↑ Miller, C. P., & Bohnhoff, M. (1963). Changes in the mouse's enteric microflora associated with enhanced susceptibility to Salmonella infection following streptomycin treatment. The Journal of infectious diseases, 59-66.
- ↑ Kinnebrew, M. A., Ubeda, C., Zenewicz, L. A., Smith, N., Flavell, R. A., & Pamer, E. G. (2010). Bacterial flagellin stimulates toll-like receptor 5—dependent defense against vancomycin-resistant Enterococcus infection. The Journal of infectious diseases, 201(4), 534-543.
- ↑ de Gunzburg, J., Ghozlane, A., Ducher, A., Le Chatelier, E., Duval, X., Ruppé, E., ... & Chachaty, E. (2018). Protection of the human gut microbiome from antibiotics. The Journal of infectious diseases, 217(4), 628-636.
- ↑ Meek, R. W., Vyas, H., & Piddock, L. J. (2015). Nonmedical Uses of Antibiotics: Time to Restrict Their Use?. PLoS biology, 13(10), e1002266. https://doi.org/10.1371/journal.pbio.1002266
Edited by Zachary Baker, student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2020, Kenyon College.
