Desulfatibacillum alkenivorans
Desulfatibacillum alkenivorans AK-01
Description and Significance
Groundwater serves as water for more than 50% of people living in North America therefore a significant public resource. To date, major contamination of groundwater in North America are due to the release and use of chlorinated ethenes by industry. Examples of such toxic compounds are perchloroethene (PCE), trichloroethene (TCE). Carbon tetrachloride (CT) is also a major groundwater pollutant [4]. These compounds were widely used as solvents for dry cleaning and in textile manufacturing. They are sufficiently water soluble and can travel through soil where they reach the groundwater. The relative high concentration of them here can be harmful [6]. Ground water is also contaminated by pollutants that are not highly toxic, but can be utilized or modified by microorganisms to become more toxic. For instance over-fertilization in agriculture leads to an increased nitrate concentration which i.e. can cause the Blue Baby syndrome. This is seen in infants younger than six month old who rely on bacteria to digest their food. Some of these bacteria also convert nitrate, a component of fertilizer, to nitrite. In the blood nitrite reacts with hemoglobin interfering with its ability to carry oxygen. The babies show sign of suffocation and gets a bluish skin [2].
Microbial metabolism of groundwater pollutants
Co-metabolism and degradation of TCE
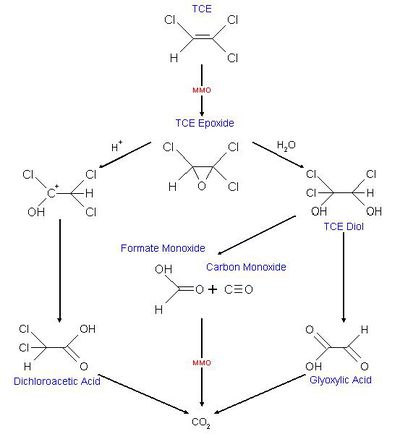
Some dehalorespiring organisms are capable of degrading PCE, TCE and CT into non-toxic compounds. Degradation of PCE is only known to happen through reductive dechlorination and only under anaerobic condition. TCE is, unlike PCE, able to be degraded under aerobic conditions. This can happen through cometabolism. In co-metabolism a compound is transformed by an organism that doesn’t use the compound as an energy or carbon source and reducing power is not provided. The organism relies on another compound to serve as an energy and carbon source [3] . Methanotrophic organisms grow on methane as a primary substrate and oxygen but some are also able to degrade TCE as a secondary substrate. This is because of nonspecific enzymatic activity of enzymes (methane monooxygenase, MMO) involved in degradation of the primary substrate. The degradation of TCE serves no beneficial purpose for these organisms. It generates an epoxide(cf. figure 1) which is transported out of the cell and here other heterotrophic organisms bring about the transformation into non-toxic compounds resulting in the formation of CO2. Several factors inhibit the aerobic degradation of TCE here among the concentration of contamination, the pH and the temperature. Because both TCE and methane bind to the same site in MMO competition between growth substrate and non-growth substrate also seems to limit degradation of TCE [3].
Dehalogenation
The bacterium Pseudomonas stutzeri strain KC can dehalogenate CT into carbon dioxide and chlorine without producing the toxic intermediate chloroform (CCl3H). This bacterium is originally isolated from an aquifer in Seal Beach in California. It is dependent on anaerobic conditions and in iron-limited media this bacterium produces and secretes a chelator called pyridine-2,6 (bis)thiocarboxylate (PDTC cf. figure 2.) [5]. When PDTC is in contact with a broad range of cell components it turns into a reduced form (the iron in the complex is reduced) and this is essential for its extracellular activity. PDCT has to be in a complex with copper in order for the fast turnover rate of CT into CO2. This complex functions both as a reactant and a catalyst in the reaction. When Pseudomonas stutzeri is in environments were nitrate is present as the electron acceptor a more rapid production of PDTC is observed [6].
Denitrification
In many agricultural areas in North America the nitrate concentrations exceed the standards. Denitrifying organisms are capable of using nitrate or nitrite as terminal electron acceptors thereby removing the excess of nitrogen from the environment. The organism Methylomirabilis oxyfera is an example of such an organism. This denitrifying bacterium is special in that it doesn’t have the gene encoding nitrous oxide reductase, the protein that converts N2O to N2. Instead they harbor an operon which encodes the complete methane monooxygenase complex. This enables it to oxide methane in an aerobic pathway [1]. The mechanism takes advances of the oxidation of methane to drive denitrification. They do so by producing oxygen from nitrite via nitrite oxide (thereby bypassing the intermediate nitrous oxide) and then use this oxygen to oxide methane in an anaerobic environment. This is called nitrite dependent anaerobic methane oxidation. The overall redox reaction is 3CH4 + 8NO2- + 8H+ -> 3CO2 + 4N2 + 10 H2O. In this way the organism uses the potent greenhouse gas methane and reduces nitrite thereby contributing to the removal of excess N-compounds in groundwater [1].
Future Applications
Organisms capable of coupling alkane degradation with sulfate reduction play a key role in sites contaminated with hydrocarbons, as these sites typically become anoxic over time due to aerobic respiration and decomposition [4]. Additionally, sulfate is relatively abundant in marine waters compared to nitrate and iron, which typically exist at low concentrations [9]. In one study, analysis of sediment polluted with petroleum after 502 days showed strains most closely related to AK-01 were the most abundant [9], further emphasizing possible bio-remedial applications of D. alkenivorans AK-01 in the future. It has been experimentally shown that AK-01 can interact with methanogens to degrade alkanes syntrophically. This mechanism could be applied within the oil industry, as the current techniques used to retrieve fossil fuels are only able to extract 40% of the available resource (8). The residual oil could be used to harvest methane gas through syntrophic degradation as an alternative energy source [8]. Current experiments expect a yield of 3 mmol methane gas per gram of residual oil. Taking into account the number of U.S. oil reservoirs, syntrophic alkane degradation could provide 17% of the natural gas consumed in the United States [8].
References
1. Callaghan, A., Gieg, L., Kropp, K., Suflita, J. and Young, L. “Comparison of mechanisms of alkane metabolism under sulfate-reducing conditions among two bacterial isolates and a bacterial consortium.” Appl Environ Microbiol, 2006, DOI: 10.1128/AEM02896-05
2. Callaghan, A., Wawrik, B., Ní Chadhain, S., Young, L. and Zylstra, G. “Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes.” Biochem Biophys Res Commun, 2008, DOI: 10.1016/j.bbrc.2007.11.094
3. Callaghan, A., Morris, B., Pereira, I., McInerney, M., Austin, R., Groves, J., Kukor, J., Suflita, J., Young, L., Zylstra, G. and Wawrik, B. “The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation.” Environ Microbiol, 2012, DOI: 10.1111/j.1462-2920.2011.02516.x
4. Canfield, D., Jorgensen, B., Fossing, H., Glud, R., Gundersen, J., Ramsing, N., Thamdrup, B., Hansen, J., Nielsen, L. and Hall, P. “Pathways of organic carbon oxidation in three continental margin sediments.” Mar Geol, 1993, DOI: 10.1016/0025-3227(93)90147-N
5. Cravo-Laureau, C., Matheron, R., Cayol, J., Joulian, C. and Hirschler-Réa, A. “Desulfatibacillum aliphaticivorans gen. nov., sp. nov., an n-alkane- and n-alkene-degrading, sulfate-reducing bacterium.” Int J Syst Evol Microbiol, 2004, DOI: 10.1099/ijs.0.02717-0
6. Davidova, I., Duncan, K., Choi, O. and Suflita, J. “Desulfoglaeba alkanexedens gen. nov., sp. nov., an n-alkane-degrading, sulfate-reducing bacterium.” Int J Syst Evol Microbiol, 2006, DOI: 10.1099/ijs.0.64398-0
7. Ferry, J., Smith, P. and Wolfe, R. “Methanospirillum, new genus of methanogenic bacteria, and characterization of Methanospirillum hungatei sp. nov.” Int J Syst Evol Microbiol, 1974, Retrieved from http://ijs.sgmjournals.org/content/24/4/465
8. Gieg, L., Duncan, K. and Suflita, J. “Bioenergy Production via Microbial Conversion of Residual Oil to Natural Gas.” Appl Environ Microbiol, 2008, DOI: 10.1128/AEM.00119-08
9. Miralles, G., Grossi, V., Acquaviva, M., Duran, R., Bertrand, J. and Cuny, P. “Alkane biodegradation and dynamics of phylogenetic subgroups of sulfate-reducing bacteria in an anoxic costal marine sediment artificially contaminated with oil.” Chemosphere, 2007, DOI: 10.1016/j.chemosphere.2007.01.033
10. So, C. and Young, L. “Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes.” Appl Environ Microbiol, 1999, Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC91444/
11. So, C. and Young, L. “Initial reactions in anaerobic alkane degradation by a sulfate reducer, strain AK-01.” Appl Environ Microbiol, 1999, Retrieved from http://www.ncbi.nlm.nih.gov/pmc/articles/PMC91754/
12. So, C., Phelps, C. and Young, L. “Anaerobic transformation of alkanes to fatty acids by a sulfate-reducing bacterium, strain Hxd3.” Appl Environ Microbiol, 2003, DOI: 10.1128/AEM.69.7.3892-3900.2003
13. U.S. Department of Energy Joint Genome Institute. “Desulfatibacillum alkenivorans AK-01.” 2010, Retrieved from http://genome.jgi.doe.gov/desa1/desa1.info.html
14. Wilkes, H., Rabus, R. and Fischer, T. “Anaerobic degradation of n-hexane in a denitrifying bacterium: Further degradation of the initial intermediate (1-methylpentyl)succinate via C-skeleton rearrangement.” Arch Microbiol, 2002, DOI: 10.1007/s00203-001-0381-3