Oral microbial system
Introduction
Oral microbiome is a complex ecological system where upto 700 species of microorganisms that have been identified (1). Some of the predominant groups present in the mouth include Streptococcus, Neisseria, Veillonella, Actinomyces and other obligate anaerobes (2). These organisms maintain a mutualistic relationship with the host by preventing pathogenic species from adhering to the mucosal surface (3). Oral microflorae can cause dental plaques and are also a common cause dental caries and periodontal disease (4, 5). Oral disease in an individual can be caused due to a combination of lack of oral hygiene and factors influencing the oral microbial community structure, such as diet. (6). An understanding of the oral environment and microbial interactions leads to understanding the main causes for the onset of oral diseases.
Oral Environment
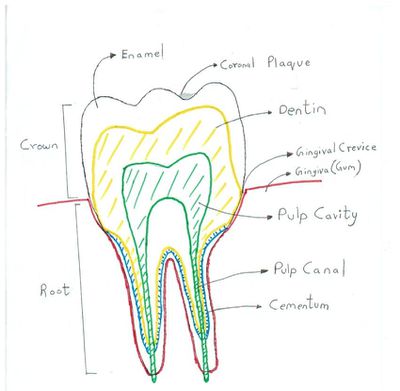
Oral microflora are most commonly found in gingival crevices, coronal plaques, tongue dorsum, buccal mucosa and saliva (6). The gingival crevices have the highest concentration of anaerobes (mainly spirochetes, vibrios & S.melaninogenicus) because it has an ideal redox potential and anoxygenic environment which helps select for the growth of these microorganisms (6). Streptococcus salivarius is found mostly in the dorsal tongue and the saliva, Streptococcus mitis is commonly found in buccal mucosa and Streptococcus sanguinis present in the teeth (8, 6).
Saliva influences the microbial ecology since it is an important source of nutrients for microorganisms and contains proteins and glycoproteins. Saliva controls microbial communities with immunoglobulins and lactoferrins which prevent growth of pathogenic microorganisms (1, 7). The saliva is also essential in the formation of an insoluble film called salivary pellicle which coats the teeth and has receptors to which adhesins on the surface of specific microorganisms can bind to (3). The physical environment (temperature, redox potential and pH) also play a strong role in selecting for the growth of oral microflora over organism that normally grow in the outside environment (6). Desquamation (shedding of epithelial cells) in the mouth ensure that only bacteria which adhere strongly are allowed to remain in the mouth and thereby limit bacterial colonization (6).
Microbial Interactions
Nutrition
Oral microflora uses substrates either derived from the diet of the host or produced by other bacteria (6). This is done by the pioneer microbial community (first microorganisms to colonize), in order to generate end products or breakdown products of metabolism, which can then be used by other microorganisms (7). Host substrates are derived from the constituents of saliva and serum-like cervicular fluid (both of which contains low amounts of glucose and proteins including albumin and proline-rich proteins) (9). Oral microorganisms are efficient at producing enzymes like proteases, lipases and glycoside hydrolases to degrade and use polymers produced by the host (6). Nutritional interrelationships between oral microorganisms in vivo have been difficult to establish given the variety of organisms collectively working together in the mouth. However, it has been suggested by Rogosa that Veilonella utilize lactic acid produced by Streptococci as it cannot ferment carbohydrates on its own (11). Similarly, Gibbons & Macdonald have shown that certain strains of Bacteroides melaninogenicus require Vitamin K produced by other bacteria for growth (10). Hence there is a symbiotic nutritional relationship between the commensal oral bacteria present in the mouth.
Signaling
Bacteria use a method of communication called quorum sensing which involves the release of special molecules used to communicate with other bacteria (2). This is particularly vital for colony growth and biofilm formation as it sets up the basis of coaggregation between different oral microflorae (See Adhesion subection).
Several microorganisms work in complex ways to coexist in the oral microbial community. One example is Streptococcus gordonii which produces H2O2 to kill other oral bacteria, but it does not have catalases to breakdown H2O2. However, another bacterial species called Actinnomyces naeslundi produce catalases which break down some of the H2O2 that S. gordonii cannot break (2). This suggests possible signalling interactions between these two organisms which allow both of them to flourish together but prevents growth competition with other bacteria. It has also been suggested by Marsh that resident microflora take active part in signalling with host epithelial cells (7). This is important as it causes the host cell to supress its inflammatory response and allows the microflora to grow. The microflora in return helps the innate host defense by preventing pathogenic species from colonizing the environment (7).
Adhesion
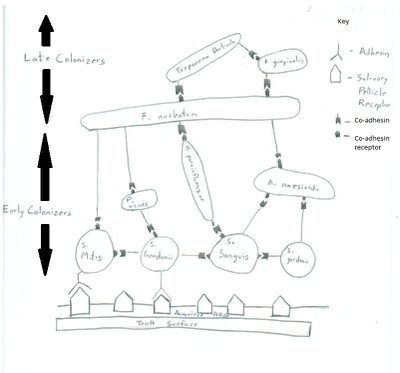
Biofilms can be formed in a variety of oral cavity surfaces including saliva coated enamel and dental surfaces. The initial attachment is done by primary colonizers (Streptococcus sanguis, Viellonella, Fusobacterium) which results in the formation of microbial monolayers (2). The adherence is initially a long ranged non-specific reversible interaction, but then followed by an irreversible short ranged adhesion-receptor interaction (7). Liljemark & Bloomquist have shown S. sanguis to have an affinity for the receptors in the salivary pellicle (3). Different species of Streptococcus produce a variety of oral adhesion including type 1 fimbrae associated proteins and antigen I/II which help the organism to attach itself to the receptors of salivary pellicle (2). With time and arrival of other late colonizers, the microbial monolayer gives rise to a multilayered bio-film as the late colonizers bind to the primary colonizers by co-adhesion (7). The late colonizers synthesize protein adhesins which cannot bind to the salivary pellicle but is efficient in binding to the receptors the primary colonizers. This mechanism where microflorae bind to each other to form biofilm is called coaggregation (12). This is important because the bacteria present in the biofilm community work together to not only outcompete pathogenic bacteria for resources (like space and nutrition) but also have mutualistic nutritional inter-relationships among themselves.
Causes of oral disease
The two main etiologies of oral diseases have been attributed to Porphyromonas gingivalis which causes periodontal diseases (like periodontitis) and Streptococcus mutans which causes dental caries (3). Both these bacteria are not ubiquitously found in the mouth because the normal flora in the oral cavity is commensal and work together to prevent pathogenic bacteria from affecting the host. Thus caries and periodontal diseases is a result of a combination poor oral hygiene and high sugar based diet which causes constant recolonization by pathogenic bacteria (3). Emperical studies by Kigure et al. have found that there is higher concentration P. gingivalis and Treponema denticola are found in the periodontal pockets of adults suffering from periodontitis (4). The proposed mechanism for periodontitis is that both these organisms have shown hemolytic activity (by possible virulence factor) and this affects the attachment of the gums with the teeth (4). In case of dental caries, Sucrose has been shown to help S. mutans colonize by helping it adhere better (6). S. mutans break sucrose down to produce glucans (extracellular polymer) which can then be used for building a biofilm which is different from biofilms produced by normal florae. This biofilm has increased porosity and allows acids to freely diffuse to the enamel and cause a pH drop, which can lead ultimately lead to dental caries (3).
The onset for both periodontitis and dental caries is a result of a shift in the normal composition of the oral flora. The mouth is in general an inhospitable place for pathogenic bacteria and thus it is interesting to know that is possible to prevent these diseases by ensuring good oral hygiene and avoiding high sugar diets.
Reference
1 Palmer Jr., R.J. , Chalmers, N.I., Rickard, A.H. and Kolenbrander, P.E. “Community Development in Bacterial Biofilms of the Oral Cavity.” Microscopy and Microanalysis, 2008, DOI: 10.1017/S1431927608088831
2 Avila, M., Ojcius, D. M. and Yilmaz, O. “The Oral Microbiota: Living with a Permanent Guest.” DNA & Cell Biology, 2009, DOI: 10.1089/dna.2009.0874
3 Liljemark, W.F. and Bloomquist, C. “Human Oral Microbial Ecology and Dental Caries and Periodontal Diseases.” Critical Reviews in Oral Biology & Medicine, 1996, DOI: 10.1177/10454411960070020601
4 Kigure, T., Saito, A., Seida, K., Yamada, S., Ishihara, K. and Okuda K. “Distribution of Porphyromonas gingivalis and Treponema denticola in human subgingival plaque at different periodontal pocket depths examined by immunohistochemical methods.” Journal of Peridontal Research, 1995, DOI: 10.1111/j.1600-0765.1995.tb01284.x
5 Sbordone, L. and Bortolaia, C. “Oral microbial biofilms and plaque-related diseases: microbial communities and their role in the shift from oral health to disease.” Clinical Oral Investigations, 2003, DOI: 10.1007/s00784-003-0236-1
6 Gibbons, R. J. and Houte, J. V. “Bacterial Adherence in Oral Microbial Ecology.” Annual Review of Microbiology, 1975, DOI: 10.1146/annurev.mi.29.100175.000315 7 Marsh, P. D. “Role of the Oral Microflora in Health.” Microbial Ecology in Health and Disease, 2000, DOI:10.1080/089106000750051800
8 Socransky, S.S. and Manganiello, S.D. “The Oral Microbiota of Man From Birth to Senility.” Journal of Periodontology, 1971, DOI 10.1902/jop.1971.42.8.485
9 Scannapieco, F.A. “Saliva-Bacterium Interactions in Oral Microbial Ecology.” Critical Reviews in Oral Biology & Medicine, 1994, doi: 10.1177/10454411940050030201
10 Gibbons, R. J. and Macdonald, J.B. “Hemin and vitamin K compounds as required factors for the cultivation of certain strains of Bacteroides melaninogenicus.” Journal of Bacteriology, 1960.
11 Rogosa, M. “The Genus Veillonella I. General Cultural, Ecological, and Biochemical Considerations.” Journal of Bacteriology, 1964.
12 Kolenbrander, P. E. and London, J. “Adhere Today, Here Tomorrow: Oral Bacterial Adherence.” Journal of Bacteriology, 1993.