Membrane proteins as mechanisms of immune system evasion in Trypanosoma brucei
Introduction
Of all defined parasitic human pathogens, the transmitter of Trypanosomiasis (commonly known as African Sleeping Sickness) has a truly fascinating relationship to its hosts. As heteroxenous organisms, Trypanosoma parasites require more than one host to successfully complete their life cycles. Specifically, these flagellate protozoa are unicellular and carry out part of their growth inside the gut of the tse tse fly prior to transmission into the human bloodstream. The mechanisms through which Trypanosoma species evade the human immune system are complex and not completely defined. Much research is concerned with studying the exact modes these unique organisms have for escaping antibody detection and wreaking havoc on their hosts’ health. Perhaps the most important technique Trypanosoma species have is the ability to manipulate their membrane receptor proteins at a more rapid speed than human immune systems can assemble appropriate antibodies. Inadequacy of current treatments has spurred on research to describe this phenomenon in greater detail and discover more effective means of understanding the many facets of African sleeping sickness.
Various species of the genus Trypanosoma confer disease to humans. The most prevalent and thus most widely studied is T. brucei, a eukaryotic flagellate. The parasite is typical in that it contains fully functional mitochondria and other membrane-bound organelles. The mitochondria of the trypanosome include kinetoplasts, disk-shaped networks of condensed circular DNA containing many copies of the mitochondrial genome.
Disease Origins and Distribution
Two morphologically indistinguishable subspecies of Trypanosoma brucei have been described to cause diverse disease patterns in humans: T. b. rhodesiense (East African sleeping sickness) and T. b. gambiense (West African sleeping sickness). The two strains are chiefly found in different regions of Africa and currently no data exists to support any overlap in distribution. Predominantly found in regions of eastern and southeastern Africa, T. b. rhodesiense infections are the most prevalent in Tanzania, Uganda, Malawi, and Zambia (CDC). Wild animals have been found to be the most effective reservoirs of these trypanosomes, capable of infecting local populations as well as sporadically transmitting to hunters and game park visitors. As a result, approximately one case per year is diagnosed in the United States (CDC). While a few hundred incidences of this form of Trypanosomiasis are reported annually, the majority of sleeping sickness cases in African stem from T. b. gambiense, found primarily in central Africa and areas of West Africa (CDC). Trypanosomiasis is most common to the poorest rural populations of some of the least developed Central African countries (Berriman et al 2005). Of the thousands of cases reported to the World Health Organization (WHO) each year, more than 95% originate from Democratic Republic of Congo, Angola, Sudan, Central African Republic, Chad and northern Uganda (Center for Disease Control and Prevention, CDC).
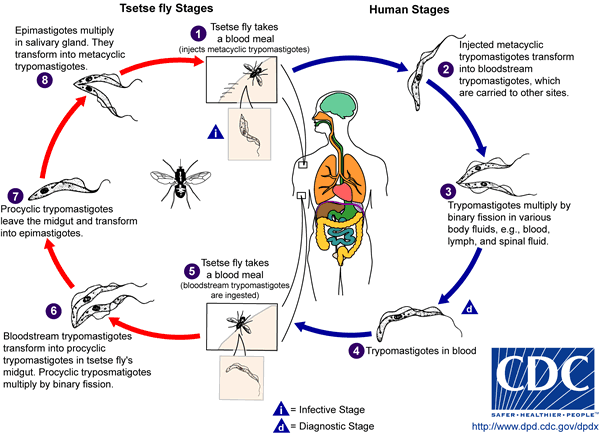
Mechanism of Infection
Include some current research in each topic, with at least one figure showing data.
Symptoms, Diagnosis, and Treatment
some stuff
Protein Cycling
talk about proteins
Drug Interactions
drugs
Future Research
further description of mechanism of action (current data)
Conclusion
Overall paper length should be 3,000 words, with at least 3 figures.
References
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.