Predation by Myxococcus xanthus
Introduction
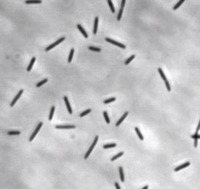
Myxococcus xanthus is a gram-negative, rod-shaped bacterium (see Figure 1), prevalent in soil environments, that exhibits extensive social behaviour [1]. When nutrients are scarce, bacteria aggregate into fruiting bodies that form spores. When adequate food is present in the environment, however, M. xanthus cells collectively swarm and prey upon other organisms. This mechanism of predation is a highly organized system involving sensing, moving towards, and lysing prey [2].
Sensing Prey
Signal Transduction
Myxococcus xanthus uses a frz signal transduction system to control its directed movement towards prey [3]. This frz system is similar to the Che chemotaxis system in E. coli, and frz gene mutants exhibit similar characteristics to well-known che gene mutants in the E. coli system. In M. xanthus, however, chemotactic activity is controlled by reversing direction of gliding motility, rather than by switching direction of flagellar rotation [4].
Direct Contact
M. xanthus cells cannot sense prey colonies until direct cell-cell contact is made. There is no recognition of prey cells even at very short distances, but when contact is made, the M. xanthus cells began to alter their behaviour. The frz signal transduction system is also responsible for keeping the M. xanthus cells in the vicinity of their prey after contact has been made and feeding is underway. When cells start moving away from the source, the frz system senses this and induces a reverse in direction to keep them in contact with the prey colony [3].
Motility During Predation
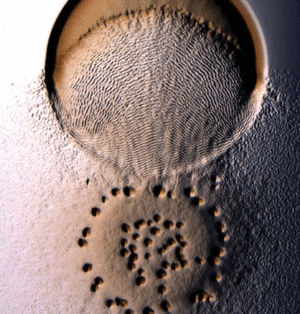
M. xanthus uses what is referred to as the “wolf pack” approach to predation [5]. Cells usually exist as biofilms consisting of a layered arrangement. Once prey is sensed, the cells cluster into organized groups known as swarms, which then invade prey colonies [2].
Predatory Rippling
Myxococcus xanthus swarms utilise a behaviour known as rippling during predation. It is induced when M. xanthus makes contact with prey or other food sources. Rippling is also observed during fruiting body formation, but research by Berleman et al. [6] demonstrates that it is generally a predatory mechanism and is necessary for efficient predation. During rippling, cells accumulate in aggregates containing ridges and troughs, and move in a manner similar to water-like ripples [7]. These ripples were coined “accordion waves” because they reflect off each other when they collide, causing each wave crest to oscillate with no net displacement. This is due to intercellular signalling during wave collision that causes changes in direction in all the cells of the swarm. [8]. Rippling behaviour has physiological benefits during predation because it retains M. xanthus cells in the area of their prey for a longer time, by decreasing random drift and increasing organized alignment [9].
Reversal Rate
Directed movement of Myxococcus xanthus relies on the reversal of direction. M. xanthus cells reverse direction on average every 7 minutes. Net movement results when the intervals between the reversals vary [4]. During predatory rippling, this allows the swarm of cells to increase its expansion in the direction of prey, because more of their potential velocity is oriented towards the prey as the reversal frequency decreases. [10]. Varying reversal rate of individual cells and therefore of the swarm is critical for movement in a particular direction.
Killing and Digestion of Prey
During predation, Myxococcus xanthus feeds by causing lysis of prey cell envelopes, and then extracting cytoplasmic contents to digest for carbon, nitrogen, and energy [11]. It is capable of killing prey organisms by collectively secreting both antibiotics and lytic enzymes. The lytic enzymes produced include cell wall cleaving enzymes such as proteases, peptidases, nucleases, and lipases [12]. Some of the antibiotics produced are bactericidal, and some only halt growth of prey organisms making them easier to lyse using other compounds [13].
Symbiosis
Ignicoccus hospitalis is the only member of the genus Ignicoccus that has been shown to have an extensive symbiotic relationship with another organism.
Ignicoccus hospitalis has been shown to engage in symbiosis with Nanoarchaeum equitans . Nanoarchaeum equitans is a very small coccoid species with a cell diameter of 0.4 µm[9] . Genome analysis has provided much of the known information about this species.
To further complicate the symbiotic relationship between both species, it’s been observed that the presence of Nanoarchaeum equitans on the surface of Ignicoccus hospitalis somehow inhibits the cell replication of Ignicoccus hospitalis . How or why this occurs has not yet been elucidated[3] .
Nanoarchaeum equitans
Nanoarchaeum equitans has the smallest non-viral genome ever sequenced at 491kb[9] . Analysis of the genome sequence indicates that 95% of the predicted proteins and stable RNA molecules are somehow involved in repair and replication of the cell and its genome[3] .
Analysis of the genome also showed that Nanoarchaeum equitans lacks nearly all genes known to be required in amino acid, nucleotide, cofactor and lipid metabolism. This is partially supported by the evidence that Nanoarchaeum equitans has been shown to derive its cell membrane from its host Ignicoccus hospitalis cell membrane. The direct contact observed between Nanoarchaeum equitans and Ignicoccus hospitalis is hypothesized to form a pore between the two organisms in order to exchange metabolites or substrates (likely from Ignicoccus hospitalis towards Nanoarchaeum equitans due to the parasitic relationship). The exchange of periplasmic vesicles is not thought to be involved in metabolite or substrate exchange despite the presence of vesicles in the periplasm of Ignicoccus hospitalis .
These analyses of the Nanoarchaeum equitans genome support the fact of the extensive symbiotic relationship between Nanoarchaeum equitans and Ignicoccus hospitalis. However, it has not yet been proven that it is a strictly parasitic relationship and further research may prove that there is a commensal relationship between the two species.
References
(1) Reichenbach H. 1999. The ecology of myxobacteria. Environmental Microbiology. 1: 15-20.
(2) Zusman DR, Scott AE, Zhaomin Y, Kirby JR. 2007. Chemosensory pathways, motility, and development in Myxococcus xanthus. Nature Reviews. 5:862-872.
(3) Giannone R.J., Heimerl T., Hettich R.L., Huber H., Karpinets T., Keller M., Kueper U., Podar M. and Rachel R. “Proteomic Characterization of Cellular and Molecular Processes that Enable the Nanoarchaeum equitans- Ignicoccus hospitalis Relationship.” PLoS ONE, 2011, Volume 6, Issue 8.
(4) Eisenreich W., Gallenberger M., Huber H., Jahn U., Junglas B., Paper W., Rachel R. and Stetter K.O. “Nanoarchaeum equitans and Ignicoccus hospitalis: New Insights into a Unique, Intimate Association of Two Archaea.” Journal of Bacteriology, 2008, DOI: 10.1128/JB.01731-07.
(5) Grosjean E., Huber H., Jahn U., Sturt H, and Summons R. “Composition of the lipids of Nanoarchaeum equitans and their origin from its host Ignicoccus sp. strain KIN4/I.” Arch Microbiol, 2004, DOI: 10.1007/s00203-004-0725-x.
(6) Briegel A., Burghardt T., Huber H., Junglas B., Rachel R., Walther P. and Wirth R. “Ignicoccus hospitalis and Nanoarchaeum equitans: ultrastructure, cell–cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography.” Arch Microbiol, 2008, DOI 10.1007/s00203-008-0402-6.
(7) Burghardt T., Huber H., Junglas B., Naether D.J. and Rachel R. “The dominating outer membrane protein of the hyperthermophilic Archaeum Ignicoccus hospitalis: a novel pore-forming complex.” Molecular Microbiology, 2007, Volume 63.
(8) Berg I.A., Eisenreich W., Eylert E., Fuchs G., Gallenberger M., Huber H.,Jahn U. and Kockelkorn D. “A dicarboxylate/4-hydroxybutyrate autotrophic carbon assimilation cycle in the hyperthermophilic Archaeum Ignicoccus hospitalis.” PNAS, 2008, Volume 105, issue 22.
(9) Brochier C., Gribaldo S., Zivanovic Y., Confalonieri F. and Forterre P. “Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales?” Genome Biology 2005, DOI:10.1186/gb-2005-6-5-r42.
(10) Huber H., Rachel R., Riehl S. and Wyschkony I. “The ultrastructure of Ignicoccus: Evidence for a novel outer membrane and for intracellular vesicle budding in an archaeon.” Archaea, 2002, Volume 1.