Bacterial Transcription Factors against Reactive Oxygen Species: Difference between revisions
| Line 20: | Line 20: | ||
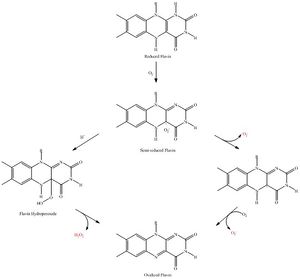
<br> Bacteria live in a world full of toxic reactive oxygen species (ROS), including hydrogen peroxide (H<sub>2</sub>O<sub>2</sub>), superoxide (O<sub>2</sub><sup>-</sup>), and hydroxyl radicals. ROS can cause severe damage to all kinds of cell components, including nucleic acids, lipids and proteins (Halliwell, 2006). These highly reactive species are generated endogenously by accidental autoxidation of flavoproteins, a group of non-respiratory redox enzymes (Figure 1) (Seaver and Imlay, 2004; Korshunov and Imlay, 2010). Thus, aerobic bacteria have evolved to maintain high concentration of enzymes that keep intracellular ROS below a lethal level. However, various conditions are known to increase these species above such threshold, inflicting oxidative stress to the bacteria, a phenomenon called “oxidative burst” (Apostol et al., 1989). Therefore, bacteria have developed defensive systems to deal with such situations. One mechanism is ROS inducible transcriptional factors that regulate gene expression of a variety of enzymes to respond to the oxidative stress. Three redox-sensitive transcription factors in bacteria have been intensively studied: OxyR and PerR against H<sub>2</sub>O<sub>2</sub>, and SoxR against O<sub>2</sub><sup>-</sup> (Reviewed by Imlay, 2015). | <br> Bacteria live in a world full of toxic reactive oxygen species (ROS), including hydrogen peroxide (H<sub>2</sub>O<sub>2</sub>), superoxide (O<sub>2</sub><sup>-</sup>), and hydroxyl radicals. ROS can cause severe damage to all kinds of cell components, including nucleic acids, lipids and proteins (Halliwell, 2006). These highly reactive species are generated endogenously by accidental autoxidation of flavoproteins, a group of non-respiratory redox enzymes (Figure 1) (Seaver and Imlay, 2004; Korshunov and Imlay, 2010). Thus, aerobic bacteria have evolved to maintain high concentration of enzymes that keep intracellular ROS below a lethal level. However, various conditions are known to increase these species above such threshold, inflicting oxidative stress to the bacteria, a phenomenon called “oxidative burst” (Apostol et al., 1989). Therefore, bacteria have developed defensive systems to deal with such situations. One mechanism is ROS inducible transcriptional factors that regulate gene expression of a variety of enzymes to respond to the oxidative stress. Three redox-sensitive transcription factors in bacteria have been intensively studied: OxyR and PerR against H<sub>2</sub>O<sub>2</sub>, and SoxR against O<sub>2</sub><sup>-</sup> (Reviewed by Imlay, 2015). | ||
<br> OxyR is induced by H<sub>2</sub>O<sub>2</sub> and activated by reversible disulfide bond formation in its active site (Choi et al., 2001). OxyR is widely distributed in Gram-negative bacteria, such as E. coli. The binding of OxyR to DNA generally turns on the gene expression of a large range of oxidative stress defensive enzymes (Zheng et al., 1998). Recent research also has shown that Gram-negative bacteria Vibrio vulnificus, facultative aerobes, contain two OxyR-type regulators with similar sensory mechanism but different sensitivity in order to fine-tune the expression of genes against H<sub>2</sub>O<sub>2</sub>pressure (Kim et al., 2014). | <br> OxyR is induced by H<sub>2</sub>O<sub>2</sub> and activated by reversible disulfide bond formation in its active site (Choi et al., 2001). OxyR is widely distributed in Gram-negative bacteria, such as E. coli. The binding of OxyR to DNA generally turns on the gene expression of a large range of oxidative stress defensive enzymes (Zheng et al., 1998). Recent research also has shown that Gram-negative bacteria Vibrio vulnificus, facultative aerobes, contain two OxyR-type regulators with similar sensory mechanism but different sensitivity in order to fine-tune the expression of genes against H<sub>2</sub>O<sub>2</sub> pressure (Kim et al., 2014). | ||
<br> PerR is the equivalent H<sub>2</sub>O<sub>2</sub>sensor protein in many Gram-positive bacteria, such as Staphylococcus aureus and Bacillus subtilis. PerR is activated by oxidation of its regulatory metal, either | <br> PerR is the equivalent H<sub>2</sub>O<sub>2</sub>sensor protein in many Gram-positive bacteria, such as Staphylococcus aureus and Bacillus subtilis. PerR is activated by oxidation of its regulatory metal, either Fe<sup>2+</sup> or Mn<sup>2+</sup>. The binding of PerR to DNA generally serves as down-regulates the expression of target genes (Li and Helmann, 2014; Ji et al., 2015). | ||
<br> SoxR is conserved widely in both proteobacteria and actinobacteria with an O<sub>2</sub><sup>-</sup> sensitive metal complex in its active site. The binding of SoxR to DNA induces the transcription of a secondary transcription factor, SoxS, which activates a group of enzymes suppressing the level of O<sub>2</sub><sup>-</sup> (Greenbert et al., 1990; Tsaneva and Weiss, 1990; Blanchard et al., 2007). | <br> SoxR is conserved widely in both proteobacteria and actinobacteria with an O<sub>2</sub><sup>-</sup> sensitive metal complex in its active site. The binding of SoxR to DNA induces the transcription of a secondary transcription factor, SoxS, which activates a group of enzymes suppressing the level of O<sub>2</sub><sup>-</sup> (Greenbert et al., 1990; Tsaneva and Weiss, 1990; Blanchard et al., 2007). | ||
Revision as of 17:38, 22 April 2016
Overview
By Jiayu Chen
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki.
The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Introduce the topic of your paper. What is your research question? What experiments have addressed your question? Applications for medicine and/or environment?
Bacteria live in a world full of toxic reactive oxygen species (ROS), including hydrogen peroxide (H2O2), superoxide (O2-), and hydroxyl radicals. ROS can cause severe damage to all kinds of cell components, including nucleic acids, lipids and proteins (Halliwell, 2006). These highly reactive species are generated endogenously by accidental autoxidation of flavoproteins, a group of non-respiratory redox enzymes (Figure 1) (Seaver and Imlay, 2004; Korshunov and Imlay, 2010). Thus, aerobic bacteria have evolved to maintain high concentration of enzymes that keep intracellular ROS below a lethal level. However, various conditions are known to increase these species above such threshold, inflicting oxidative stress to the bacteria, a phenomenon called “oxidative burst” (Apostol et al., 1989). Therefore, bacteria have developed defensive systems to deal with such situations. One mechanism is ROS inducible transcriptional factors that regulate gene expression of a variety of enzymes to respond to the oxidative stress. Three redox-sensitive transcription factors in bacteria have been intensively studied: OxyR and PerR against H2O2, and SoxR against O2- (Reviewed by Imlay, 2015).
OxyR is induced by H2O2 and activated by reversible disulfide bond formation in its active site (Choi et al., 2001). OxyR is widely distributed in Gram-negative bacteria, such as E. coli. The binding of OxyR to DNA generally turns on the gene expression of a large range of oxidative stress defensive enzymes (Zheng et al., 1998). Recent research also has shown that Gram-negative bacteria Vibrio vulnificus, facultative aerobes, contain two OxyR-type regulators with similar sensory mechanism but different sensitivity in order to fine-tune the expression of genes against H2O2 pressure (Kim et al., 2014).
PerR is the equivalent H2O2sensor protein in many Gram-positive bacteria, such as Staphylococcus aureus and Bacillus subtilis. PerR is activated by oxidation of its regulatory metal, either Fe2+ or Mn2+. The binding of PerR to DNA generally serves as down-regulates the expression of target genes (Li and Helmann, 2014; Ji et al., 2015).
SoxR is conserved widely in both proteobacteria and actinobacteria with an O2- sensitive metal complex in its active site. The binding of SoxR to DNA induces the transcription of a secondary transcription factor, SoxS, which activates a group of enzymes suppressing the level of O2- (Greenbert et al., 1990; Tsaneva and Weiss, 1990; Blanchard et al., 2007).
Sample citations: [1]
[2]
A citation code consists of a hyperlinked reference within "ref" begin and end codes.
Section 1
Include some current research, with at least one figure showing data.
Every point of information REQUIRES CITATION using the citation tool shown above.
Section 2
Include some current research, with at least one figure showing data.
Section 3
Include some current research, with at least one figure showing data.
Section 4
Conclusion
References
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2016, Kenyon College.