Bacterial Transcription Factors against Reactive Oxygen Species: Difference between revisions
No edit summary |
|||
| Line 6: | Line 6: | ||
<br> OxyR is induced by H<sub>2</sub>O<sub>2</sub> and activated by reversible disulfide bond formation in its active site. <ref>[http://www.sciencedirect.com/science/article/pii/S0092867401003002 Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.]</ref> OxyR is widely distributed in Gram-negative bacteria, such as <i>E. coli</i>. The binding of OxyR to DNA generally turns on the gene expression of a large range of oxidative stress defense enzymes. <ref>[http://jb.asm.org/content/183/15/4562.full Zheng et al. (1998): DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. Journal of Bacteriology 183(15): 4562-4570.]</ref> Recent research also has shown that Gram-negative bacteria <i>Vibrio vulnificus</i>, facultative aerobes, contain two OxyR-type regulators with similar sensory mechanism but different sensitivity in order to fine-tune the expression of genes against H<sub>2</sub>O<sub>2</sub> pressure.<ref>[http://onlinelibrary.wiley.com/doi/10.1111/mmi.12712/full Kim et al. (2014): Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in <i>Vibrio vulnificus</i>. Molecular Microbiology 93(5):992–1009.]</ref> | <br> OxyR is induced by H<sub>2</sub>O<sub>2</sub> and activated by reversible disulfide bond formation in its active site. <ref>[http://www.sciencedirect.com/science/article/pii/S0092867401003002 Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.]</ref> OxyR is widely distributed in Gram-negative bacteria, such as <i>E. coli</i>. The binding of OxyR to DNA generally turns on the gene expression of a large range of oxidative stress defense enzymes. <ref>[http://jb.asm.org/content/183/15/4562.full Zheng et al. (1998): DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. Journal of Bacteriology 183(15): 4562-4570.]</ref> Recent research also has shown that Gram-negative bacteria <i>Vibrio vulnificus</i>, facultative aerobes, contain two OxyR-type regulators with similar sensory mechanism but different sensitivity in order to fine-tune the expression of genes against H<sub>2</sub>O<sub>2</sub> pressure.<ref>[http://onlinelibrary.wiley.com/doi/10.1111/mmi.12712/full Kim et al. (2014): Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in <i>Vibrio vulnificus</i>. Molecular Microbiology 93(5):992–1009.]</ref> | ||
<br> PerR is the equivalent H<sub>2</sub>O<sub>2</sub> sensor protein in many Gram-positive bacteria, such as <i>Staphylococcus aureus</i> and <i>Bacillus subtilis</i>. When PerR is in its reduced form, its binding to DNA generally serves as an inhibitor of the expression of target genes. PerR is activated by oxidation of its regulatory metal, either Fe<sup>2+</sup> or Mn<sup>2+</sup>, which leads to conformational change in the protein and dissociation from DNA, thus relieving the inhibition and initiating transcription of H<sub>2</sub>O<sub>2</sub> defensive genes. <ref>[http://www.jbc.org/content/290/33/20374.full Ji et al.(2015): <i>Staphylococcus aureus</i> PerR Is a Hypersensitive Hydrogen Peroxide Sensor using Iron-mediated Histidine Oxidation. The Journal of Biological Chemistry 290:20374-20386.]</ref> <ref>[http://www.nature.com/nature/journal/v440/n7082/full/nature04537.html Lee and Helmann (2006): The PerR transcription factor senses H<sub>2</sub>O<sub>2</sub> by metal-catalysed histidine oxidation. Nature 440:363-367.]</ref> | <br> PerR is the equivalent H<sub>2</sub>O<sub>2</sub> sensor protein in many Gram-positive bacteria, such as <i>Staphylococcus aureus</i> and <i>Bacillus subtilis</i>. When PerR is in its reduced form, its binding to DNA generally serves as an inhibitor of the expression of target genes. PerR is activated by oxidation of its regulatory metal, either Fe<sup>2+</sup> or Mn<sup>2+</sup>, which leads to conformational change in the protein and dissociation from DNA, thus relieving the inhibition and initiating transcription of H<sub>2</sub>O<sub>2</sub> defensive genes. <ref>[http://www.jbc.org/content/290/33/20374.full Ji et al.(2015): <i>Staphylococcus aureus</i> PerR Is a Hypersensitive Hydrogen Peroxide Sensor using Iron-mediated Histidine Oxidation. The Journal of Biological Chemistry 290:20374-20386.]</ref> <ref>[http://www.nature.com/nature/journal/v440/n7082/full/nature04537.html Lee and Helmann (2006): The PerR transcription factor senses H<sub>2</sub>O<sub>2</sub> by metal-catalysed histidine oxidation. Nature 440:363-367.]</ref> | ||
<br> SoxR is conserved widely in both proteobacteria and actinobacteria with an O<sub>2</sub><sup>-</sup> sensitive metal complex in its active site. The binding of SoxR to DNA induces the transcription of a secondary transcription factor, SoxS, which activates a group of enzymes suppressing the level of O<sub>2</sub><sup>-</sup>. <ref>[http://www.pnas.org/content/87/16/6181.full.pdf Greenberg et al. (1990): Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in <i>Escherichia coli</i>. | <br> SoxR is conserved widely in both proteobacteria and actinobacteria with an O<sub>2</sub><sup>-</sup> sensitive metal complex in its active site. The binding of SoxR to DNA induces the transcription of a secondary transcription factor, SoxS, which activates a group of enzymes suppressing the level of O<sub>2</sub><sup>-</sup>. <ref>[http://www.pnas.org/content/87/16/6181.full.pdf Greenberg et al. (1990): Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in <i>Escherichia coli</i>. PNAS 87: 6181-6185.]</ref> <ref>[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC213242/ Tsaneva and Weiss, (1990): <i>soxR</i>, a locus governing a superoxide response regulon in <i>Escherichia coli</i> K-12. Journal of Bacteriology 172(8):4197-205.]</ref> <ref>[http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0001186 Blanchard et al. (2007): Rapid Changes in Gene Expression Dynamics in Response to Superoxide Reveal SoxRS-Dependent and Independent Transcriptional Networks. PLoS ONE 2(11): e1186.]</ref><br> | ||
==Defending against hydrogen peroxide (H<sub>2</sub>O<sub>2</sub>)== | ==Defending against hydrogen peroxide (H<sub>2</sub>O<sub>2</sub>)== | ||
| Line 12: | Line 12: | ||
Environmental H<sub>2</sub>O<sub>2</sub> is primarily generated as a metabolism by-product and as defense mechanism of plants, animals and bacteria to other microbes. <ref>[http://www.jstor.org/stable/pdf/4275862.pdf Mehdy, (1994): Active Oxygen Species in Plant Defense against Pathogens. Plant Physiology 105(2):467-472.]</ref> <ref>[http://physrev.physiology.org/content/87/1/245.short Bedard and Krause (2007): The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiological Reviews 87(1):245-313.]</ref> <ref>[http://jb.asm.org/content/186/7/2046.full.pdf Seki et al. (2004): Hydrogen Peroxide Production in <i>Streptococcus pyogenes</i>: Involvement of Lactate Oxidase and Coupling with Aerobic Utilization of Lactate. Journal of Bacteriology 186(7):2046–2051.]</ref> Because its chemical structure is similar to that of water and has no charge, H<sub>2</sub>O<sub>2</sub> can easily penetrate the membrane. Once it enters the cell, H<sub>2</sub>O<sub>2</sub> damages the cell components through the Fenton Reaction in which H<sub>2</sub>O<sub>2</sub> oxidizes Fe<sup>2+</sup> ion, generating one Fe<sup>3+</sup> ion and one hydroxyl radical. Free hydroxyl radicals can further react with lipids, nucleic acids and the disulfide bonds in proteins, which are all critical cellular components. <ref>[http://www.fasebj.org/content/1/6/441.full.pdf Machlin and Bendich (1987): Free radical tissue damage: protective role of antioxidant nutrients. The FASEB Journal 1(6):441-445.]</ref> For example, guanine is one of the most vulnerable targets to hydroxyl radical because of its small reduction potential. The resulting product is often highly mutagenic 8-hydroxyguanine, because it is able to base pair with adenine such a way that allows it to escape from the DNA mismatch detection system. <ref>[http://www.sciencedirect.com/science/article/pii/S0959440X05000151 Hogg et al. (2005): Bumps in the road: how replicative DNA polymerases see DNA damage. Current opinion in structural biology 15(1): 86-93.]</ref> The Fenton reaction also impedes the function of Fe<sup>2+</sup>-dependent enzymes that are distributed in various cellular processes.<ref>[http://science.sciencemag.org/content/240/4852/640 Imlay et al. (1988): Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240(4852): 640-642.]</ref><br> | Environmental H<sub>2</sub>O<sub>2</sub> is primarily generated as a metabolism by-product and as defense mechanism of plants, animals and bacteria to other microbes. <ref>[http://www.jstor.org/stable/pdf/4275862.pdf Mehdy, (1994): Active Oxygen Species in Plant Defense against Pathogens. Plant Physiology 105(2):467-472.]</ref> <ref>[http://physrev.physiology.org/content/87/1/245.short Bedard and Krause (2007): The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiological Reviews 87(1):245-313.]</ref> <ref>[http://jb.asm.org/content/186/7/2046.full.pdf Seki et al. (2004): Hydrogen Peroxide Production in <i>Streptococcus pyogenes</i>: Involvement of Lactate Oxidase and Coupling with Aerobic Utilization of Lactate. Journal of Bacteriology 186(7):2046–2051.]</ref> Because its chemical structure is similar to that of water and has no charge, H<sub>2</sub>O<sub>2</sub> can easily penetrate the membrane. Once it enters the cell, H<sub>2</sub>O<sub>2</sub> damages the cell components through the Fenton Reaction in which H<sub>2</sub>O<sub>2</sub> oxidizes Fe<sup>2+</sup> ion, generating one Fe<sup>3+</sup> ion and one hydroxyl radical. Free hydroxyl radicals can further react with lipids, nucleic acids and the disulfide bonds in proteins, which are all critical cellular components. <ref>[http://www.fasebj.org/content/1/6/441.full.pdf Machlin and Bendich (1987): Free radical tissue damage: protective role of antioxidant nutrients. The FASEB Journal 1(6):441-445.]</ref> For example, guanine is one of the most vulnerable targets to hydroxyl radical because of its small reduction potential. The resulting product is often highly mutagenic 8-hydroxyguanine, because it is able to base pair with adenine such a way that allows it to escape from the DNA mismatch detection system. <ref>[http://www.sciencedirect.com/science/article/pii/S0959440X05000151 Hogg et al. (2005): Bumps in the road: how replicative DNA polymerases see DNA damage. Current opinion in structural biology 15(1): 86-93.]</ref> The Fenton reaction also impedes the function of Fe<sup>2+</sup>-dependent enzymes that are distributed in various cellular processes.<ref>[http://science.sciencemag.org/content/240/4852/640 Imlay et al. (1988): Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240(4852): 640-642.]</ref><br> | ||
===<br><b>Defensive strategy for Gram-negative bacteria: OxyR</b>=== | ===<br><b>Defensive strategy for Gram-negative bacteria: OxyR</b>=== | ||
<b>H<sub>2</sub>O<sub>2</sub> sensory mechanism of OxyR</b> | |||
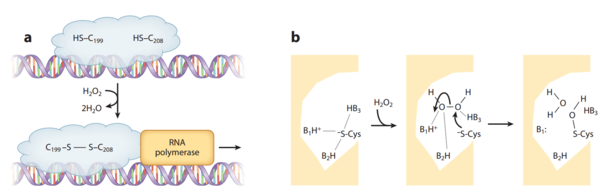
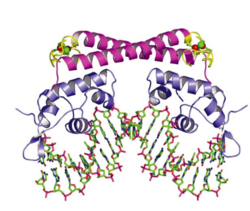
Crystal structures of OxyR in different bacteria have shown that it is a tetrameric protein. Each monomer contains a C-terminal regulatory domain and an N-terminal DNA-binding domain | Crystal structures of OxyR in different bacteria have shown that it is a tetrameric protein. Each monomer contains a C-terminal regulatory domain and an N-terminal DNA-binding domain.The redox-switch mechanism in OxyR depends on two H2O2 sensory cysteine residues (Cys-199 and Cys-208) in its C-terminal. Cys-199 residue locates in a hydrophobic pocket, where it is stabilized by hydrophobic interaction of nearby Leucine residues. Cys-199 is rapidly oxidized by H2O2 and forms in a sulfenic acid intermediate (-SOH). This intermediate is not stable in the hydrophobic pocket, and thus lead to a conformational change that flips out the oxidized Cys-199. A flexible amino acid region between these two Cysteine residues then brings Cys-199 to the proximity of the Cys-208 to form a disulfide bond. This bond formation induces OxyR conformational change in its regulatory domain, recruiting RNA polymerase and thus stimulating the expression of genes against H<sub>2</sub>O<sub>2</sub> stress.<ref>[http://www.sciencedirect.com/science/article/pii/S0092867401003002 Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.]</ref> <ref>[http://www.pnas.org/content/112/20/6443.full.pdf Jo et al. (2014): Structural details of the OxyR peroxide-sensing mechanism. PNAS 112(20):6443–6448.]</ref> Furthermore, the interaction between OxyR and RNA polymerase seems to be able to reduce the disulfide bond and makes the transcriptional regulation reversible.<ref>[http://www.sciencedirect.com/science/article/pii/S0092867401003002 Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.]</ref> [[Image:OxyR.png|thumb|600px|left|Activation of OxyR. (a) Oxidation of the H<sub>2</sub>O<sub>2</sub> sensitive Cysteine residues activate gene transcription. (b) Reaction mechanism of OxyR H<sub>2</sub>O<sub>2</sub> sensitivity ]] | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 23:01, 26 April 2016
Overview
By Jiayu Chen
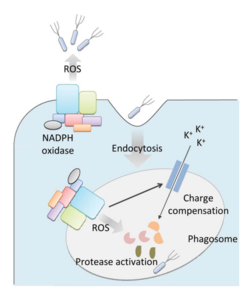
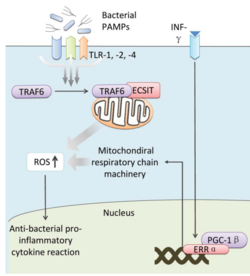
Bacteria live in a world full of toxic reactive oxygen species (ROS), including hydrogen peroxide (H2O2), superoxide (O2-), and hydroxyl radicals. ROS can cause severe damage to all kinds of cell components, including nucleic acids, lipids and proteins [1]. These highly reactive species are generated endogenously by accidental autoxidation of flavoproteins, a group of non-respiratory redox enzymes. [2] Thus, aerobic bacteria have evolved to maintain high concentration of enzymes that keep intracellular ROS below a lethal level. However, various conditions are known to increase these species above such threshold, inflicting oxidative stress to the bacteria, a phenomenon called “oxidative burst.” [3] One of the major scenario is when bacteria encounter ROS generated by the immune system of their host as a defensive weapon. In human, phagocytes including neutrophils use NADPH oxidase 2 (NOX2) complex as one of the major enzyme mediating the immune response by generating antimicrobial ROS. The resultant ROS would be secreted either outside of the cell as a trap to kill bacteria, or into phagolysosome where internalized bacteria would be degraded. Another situation that may happen during bacterial infection is the ROS generated by the mitochondria. Such defensive mechanism is mediated by many signaling molecule in the Toll pathways including several Toll-like receptors (TLRs), tumor necrosis factor receptor-associated factor 6 (TRAF6) and evolutionarily conserved signaling intermediate in Toll pathways (ECSIT).
The activation of Toll pathways resulted in increased cellular ROS as well as anti-bacterial pro-inflammatory cytokine reaction in the nucleus. Pro-inflammatory Th1 cytokine IFN-γ is also involved as a transcriptional regulator via activation of estrogen-related receptor α (ERRα) and PPARγ-coactivator-1β (PGC-1β) in the nucleus. Such interaction induces the genes that encode mitochondrial respiratory chain machinery and thus increase cellular ROS concentration. [4]
Therefore, bacteria have developed defensive systems to deal with such situations. One mechanism is ROS inducible transcriptional factors that regulate gene expression of a variety of enzymes to respond to the oxidative stress. Three redox-sensitive transcription factors in bacteria have been intensively studied: OxyR and PerR against H2O2, and SoxR against O2- [5].
OxyR is induced by H2O2 and activated by reversible disulfide bond formation in its active site. [6] OxyR is widely distributed in Gram-negative bacteria, such as E. coli. The binding of OxyR to DNA generally turns on the gene expression of a large range of oxidative stress defense enzymes. [7] Recent research also has shown that Gram-negative bacteria Vibrio vulnificus, facultative aerobes, contain two OxyR-type regulators with similar sensory mechanism but different sensitivity in order to fine-tune the expression of genes against H2O2 pressure.[8]
PerR is the equivalent H2O2 sensor protein in many Gram-positive bacteria, such as Staphylococcus aureus and Bacillus subtilis. When PerR is in its reduced form, its binding to DNA generally serves as an inhibitor of the expression of target genes. PerR is activated by oxidation of its regulatory metal, either Fe2+ or Mn2+, which leads to conformational change in the protein and dissociation from DNA, thus relieving the inhibition and initiating transcription of H2O2 defensive genes. [9] [10]
SoxR is conserved widely in both proteobacteria and actinobacteria with an O2- sensitive metal complex in its active site. The binding of SoxR to DNA induces the transcription of a secondary transcription factor, SoxS, which activates a group of enzymes suppressing the level of O2-. [11] [12] [13]
Defending against hydrogen peroxide (H2O2)
Why is H2O2 poisonous?
Environmental H2O2 is primarily generated as a metabolism by-product and as defense mechanism of plants, animals and bacteria to other microbes. [14] [15] [16] Because its chemical structure is similar to that of water and has no charge, H2O2 can easily penetrate the membrane. Once it enters the cell, H2O2 damages the cell components through the Fenton Reaction in which H2O2 oxidizes Fe2+ ion, generating one Fe3+ ion and one hydroxyl radical. Free hydroxyl radicals can further react with lipids, nucleic acids and the disulfide bonds in proteins, which are all critical cellular components. [17] For example, guanine is one of the most vulnerable targets to hydroxyl radical because of its small reduction potential. The resulting product is often highly mutagenic 8-hydroxyguanine, because it is able to base pair with adenine such a way that allows it to escape from the DNA mismatch detection system. [18] The Fenton reaction also impedes the function of Fe2+-dependent enzymes that are distributed in various cellular processes.[19]
Defensive strategy for Gram-negative bacteria: OxyR
H2O2 sensory mechanism of OxyR
Crystal structures of OxyR in different bacteria have shown that it is a tetrameric protein. Each monomer contains a C-terminal regulatory domain and an N-terminal DNA-binding domain.The redox-switch mechanism in OxyR depends on two H2O2 sensory cysteine residues (Cys-199 and Cys-208) in its C-terminal. Cys-199 residue locates in a hydrophobic pocket, where it is stabilized by hydrophobic interaction of nearby Leucine residues. Cys-199 is rapidly oxidized by H2O2 and forms in a sulfenic acid intermediate (-SOH). This intermediate is not stable in the hydrophobic pocket, and thus lead to a conformational change that flips out the oxidized Cys-199. A flexible amino acid region between these two Cysteine residues then brings Cys-199 to the proximity of the Cys-208 to form a disulfide bond. This bond formation induces OxyR conformational change in its regulatory domain, recruiting RNA polymerase and thus stimulating the expression of genes against H2O2 stress.[20] [21] Furthermore, the interaction between OxyR and RNA polymerase seems to be able to reduce the disulfide bond and makes the transcriptional regulation reversible.[22]
Section 2
Include some current research, with at least one figure showing data.
Section 3
Include some current research, with at least one figure showing data.
Section 4
Conclusion
References
- ↑ Halliwell (2006): Reactive Species and Antioxidants. Redox Biology Is a Fundamental Theme of Aerobic Life. Plant Physiology 141(2):312–322.
- ↑ Seaver and Imlay (2004): Are Respiratory Enzymes the Primary Sources of Intracellular Hydrogen Peroxide? The Journal of Biological Chemistry, 279:48742-48750.
- ↑ Apostol et al. (1989): Rapid Stimulation of an Oxidative Burst during Elicitation of Cultured Plant Cells: Role in Defense and Signal Transduction—Commentary. Plant Physiology, 90:109–116.
- ↑ Reviewed by Yang et al.(2013): Reactive oxygen species in the immune system. International Reviews of Immunology 32(3):249-70.
- ↑ Imaly, J.A. (2015): Transcription factors that defend bacteria against reactive oxygen species 2015. Annual Review of Microbiology 69:93–108.
- ↑ Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.
- ↑ Zheng et al. (1998): DNA Microarray-Mediated Transcriptional Profiling of the Escherichia coli Response to Hydrogen Peroxide. Journal of Bacteriology 183(15): 4562-4570.
- ↑ Kim et al. (2014): Distinct characteristics of OxyR2, a new OxyR-type regulator, ensuring expression of Peroxiredoxin 2 detoxifying low levels of hydrogen peroxide in Vibrio vulnificus. Molecular Microbiology 93(5):992–1009.
- ↑ Ji et al.(2015): Staphylococcus aureus PerR Is a Hypersensitive Hydrogen Peroxide Sensor using Iron-mediated Histidine Oxidation. The Journal of Biological Chemistry 290:20374-20386.
- ↑ Lee and Helmann (2006): The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature 440:363-367.
- ↑ Greenberg et al. (1990): Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. PNAS 87: 6181-6185.
- ↑ Tsaneva and Weiss, (1990): soxR, a locus governing a superoxide response regulon in Escherichia coli K-12. Journal of Bacteriology 172(8):4197-205.
- ↑ Blanchard et al. (2007): Rapid Changes in Gene Expression Dynamics in Response to Superoxide Reveal SoxRS-Dependent and Independent Transcriptional Networks. PLoS ONE 2(11): e1186.
- ↑ Mehdy, (1994): Active Oxygen Species in Plant Defense against Pathogens. Plant Physiology 105(2):467-472.
- ↑ Bedard and Krause (2007): The NOX Family of ROS-Generating NADPH Oxidases: Physiology and Pathophysiology. Physiological Reviews 87(1):245-313.
- ↑ Seki et al. (2004): Hydrogen Peroxide Production in Streptococcus pyogenes: Involvement of Lactate Oxidase and Coupling with Aerobic Utilization of Lactate. Journal of Bacteriology 186(7):2046–2051.
- ↑ Machlin and Bendich (1987): Free radical tissue damage: protective role of antioxidant nutrients. The FASEB Journal 1(6):441-445.
- ↑ Hogg et al. (2005): Bumps in the road: how replicative DNA polymerases see DNA damage. Current opinion in structural biology 15(1): 86-93.
- ↑ Imlay et al. (1988): Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240(4852): 640-642.
- ↑ Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.
- ↑ Jo et al. (2014): Structural details of the OxyR peroxide-sensing mechanism. PNAS 112(20):6443–6448.
- ↑ Choi et al. (2013): Structural Basis of the Redox Switch in the OxyR Transcription Factor. Cell 105(1):103-113.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2016, Kenyon College.