CTXφ Bacteriophage
Overview
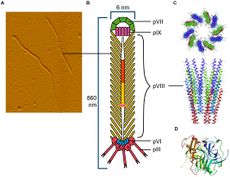
Link: https://www.frontiersin.org/articles/10.3389/fmicb.2016.00429/full
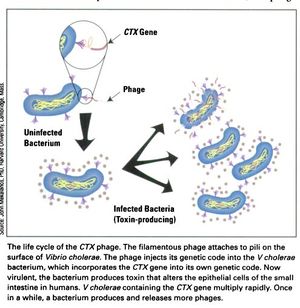
The CTXφ bacteriophage (or sometimes written as CTXphi bacteriophage) is a lysogenic, filamentous, single-stranded DNA (ssDNA) phage that is responsible for turning the previously non-infectious Vibrio cholerae into a highly pathogenic microbe that causes disease in humans.[2],[3], [4]
Genetic Material
Include some current research, with at least one image.
Infection, Replication & Lysing of Host Cell
In a process known as lysogenic phage conversion, the CTXφ bacteriophage integrates, among others, its ctxAB genes into its host, Vibrio cholerae. It is this endotoxin, called Cholera toxin or just "CT," that causes V. cholerae to switch from being nonpathogenic to highly virulent.[4]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
CT & non-CT Toxins
Include some current research, with a second image.
Conclusion
Overall text length should be at least 1,000 words (before counting references), with at least 2 images. Include at least 5 references under Reference section.
References
- ↑ Gagic, D., Ciric M., Wen W., Ng F., Rakonjac J. (2016). "Exploring the Secretomes of Microbes and Microbial Communities Using Filamentous Phage Display." Frontiers in Microbiology, 7:429. https://doi.org/10.3389/fmicb.2016.00429.
- ↑ Davis, B. M., Kimsey, H. H., Chang, W., & Waldor, M. K. (1999). "The Vibrio cholerae O139 Calcutta bacteriophage CTXφ is infectious and encodes a novel repressor." Journal of Bacteriology, 181(21), 6779-6787. https://www.frontiersin.org/articles/10.3389/fmicb.2016.00429/full
- ↑ Ochman, H., Lawrence, J. & Groisman, E. (2000). "Lateral gene transfer and the nature of bacterial innovation." Nature, 405, 299–304. https://doi.org/10.1038/35012500.
- ↑ 4.0 4.1 Boyd, E. F. (2010). "Efficiency and specificity of CTXphi chromosomal integration: dif makes all the difference." Proceedings of the National Academy of Sciences of the United States of America, 107(9), 3951–3952. https://doi.org/10.1073/pnas.1000310107
Edited by Tara Cerny, student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2019, Kenyon College.