Cauliflower mosaic virus: Difference between revisions
No edit summary |
No edit summary |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 12: | Line 12: | ||
***'''Genus''': Caulimovirus | ***'''Genus''': Caulimovirus | ||
****'''Species''': Cauliflower mosaic virus [[#References|[4]]] | ****'''Species''': Cauliflower mosaic virus [[#References|[4]]] | ||
[[Image:Cauliflower_Mosaic_Virus.jpg|thumb|300px|right|'''Fig. 1''' Cauliflower Mosaic Virus [http://www.biologydiscussion.com/plants/list-of-viruses-found-in-plants-microbiology/65790].]] | |||
==Description and Significance== | ==Description and Significance== | ||
Mosaic plant diseases are common diseases that occur during warmer months which are caused by plant mosaic viruses. These plant mosaic viruses are identified in plants based on their “mottling” and discoloration of leaves[[#References|[2]]]. Viruses may also cause leaf deformity, smaller produce and stunted growth. This could lead to plant death and a reduction of crop production which is why the study of mosaic viruses are essential to improve agriculture by both quantity and quality. | Mosaic plant diseases are common diseases that occur during warmer months which are caused by plant mosaic viruses. These plant mosaic viruses are identified in plants based on their “mottling” and discoloration of leaves[[#References|[2]]]. Viruses may also cause leaf deformity, smaller produce and stunted growth. This could lead to plant death and a reduction of crop production which is why the study of mosaic viruses are essential to improve agriculture by both quantity and quality. | ||
| Line 23: | Line 23: | ||
<br> | <br> | ||
<br> | <br> | ||
[[Image: | [[Image:CaMV_genome.jpg|thumb|300px|right|'''Fig. 2''' Cauliflower Mosaic Virus Genome[http://www.biologydiscussion.com/plants/list-of-viruses-found-in-plants-microbiology/65790].]] | ||
==Structure and Genome== | ==Structure and Genome== | ||
Cauliflower Mosaic Virus is classified as an icosahedral (20 sided) virus. This is because of the icosahedral shape of the capsid that surrounds the viral genome. This shape gives the virus 20 symmetrical triangular faces. This symmetry allows for synthesis efficiency by having the same small number of proteins being produced from a small number genes and from short chromosome sequences. This allows the largest and most complex capsid to be formed from the least amount of resources making it more efficient than a non-symmetrical capsid[[#References|[16]]]. The structure of cauliflower mosaic virus is approximately 52 nanometers in diameter and is surrounded by 420 capsid proteins arranged in triangulation T=7[[#References|[14]]]. In addition to capsid proteins, caulimoviruses are also surrounded by virus associated proteins[[#References|[5]]]. These proteins are responsible for assisting in the binding of the virus to DNA on its c-terminal end[[#References|[6]]]. | Cauliflower Mosaic Virus is classified as an icosahedral (20 sided) virus. This is because of the icosahedral shape of the capsid that surrounds the viral genome. This shape gives the virus 20 symmetrical triangular faces ('''Fig, 1'''). This symmetry allows for synthesis efficiency by having the same small number of proteins being produced from a small number genes and from short chromosome sequences. This allows the largest and most complex capsid to be formed from the least amount of resources making it more efficient than a non-symmetrical capsid[[#References|[16]]]. The structure of cauliflower mosaic virus is approximately 52 nanometers in diameter and is surrounded by 420 capsid proteins arranged in triangulation T=7[[#References|[14]]]. In addition to capsid proteins, caulimoviruses are also surrounded by virus associated proteins[[#References|[5]]]. These proteins are responsible for assisting in the binding of the virus to DNA on its c-terminal end[[#References|[6]]]. | ||
The efficiency of the icosahedral structure allows it so that the genome of CaMV does not even need to encode for more than 7 genes that already have close similarities. Genes differentiate by where their start codons are so that they have different reading frames[[#References|[16]]]. | The efficiency of the icosahedral structure allows it so that the genome of CaMV does not even need to encode for more than 7 genes that already have close similarities. Genes differentiate by where their start codons are so that they have different reading frames[[#References|[16]]]. | ||
The virion that is encapsulated in the capsid structure is made up of around 8,000 base pairs. This double stranded DNA has an open and circular shape. Also, these strands contain nicks that come from reverse transcription. These nicks are only repaired once these DNA strands enter the host when they form supercoiled molecules responsible for binding to proteins[[#References|[14]]]. | The virion that is encapsulated in the capsid structure is made up of around 8,000 base pairs. This double stranded DNA has an open and circular shape ('''Fig. 2'''). Also, these strands contain nicks that come from reverse transcription. These nicks are only repaired once these DNA strands enter the host when they form supercoiled molecules responsible for binding to proteins[[#References|[14]]]. | ||
Cauliflower mosaic virus is well known for its strong constitutive | Cauliflower mosaic virus is well known for its strong constitutive 35S promoter. This promoter runs transcription for the entire cauliflower mosaic virus genome. It is because of its efficiency, it is commonly used for studying transgenic plants in relation to gene transfer vectors. The 600 nucleotide leader sequence of the 35S promoter consists of 8 open reading frames that each possess different functions[[#References|[15]]]. There are 6 major coding regions and 2 minor coding regions. Open reading frame I functions to produce movement proteins to assist progeny viruses to pass through plasmodesmata into uninfected cells[[#References|[7]]]. Open reading frame II produces aphid transmission factors which is required for the transmission of the virus from the aphid to the plant[[#References|[8]]]. Open reading frame III, which previously had no assigned function and is a minor coding region, is believed to promote DNA binding as well as produce structural proteins. ORF IV is responsible for the production of capsid proteins that surround and protect viral genomes. ORF V produces proteins that have proteinases as well as participates in the reverse transcriptase process[[#References|[8]]]. Open reading frame VI produces trans activator proteins that promote formation of inclusion bodies. Inclusion bodies are places of viral multiplication needed to produce many progeny virions. The second minor coding region, open reading frame VII, has an unknown function but is speculated to have some function relating to targeting new viruses and bringing them to inclusion bodies (P, Ashwathi.). A very unique function of open reading frame VI that is encoded by the 19S RNA is that it can reinitiate major reading frames on the 35S RNA when this is usually only found to happen in bacteria[[#References|[15]]]. | ||
<br> | <br> | ||
<br> | <br> | ||
[[Image: | [[Image:Myszus-persicae.jpg|thumb|300px|right|'''Fig 3.''' Myszus ''persicae'' [https://www.cropscience.bayer.com/en/crop-compendium/pests-diseases-weeds/pests/myzus-persicae].]] | ||
==Ecology and Pathology== | ==Ecology and Pathology== | ||
In order for plant viruses to enter the host’s genome, it must enter via mechanical transmission. This is because cell walls are too thick for some viruses to penetrate so they rely on transmission through broken cells, animal vectors or through their own seeds[[#References|[16]]]. For Cauliflower Mosaic Virus, Aphids are the most common vector for infection. Aphids possess a needle-like mouthpiece that allows penetration into plants. Aphids then ingest the plants nutrients and sap while also leaving saliva behind in the plant. This saliva can infect the plant if the aphid contains a virus coming from a plant that was ingested beforehand[[#References|[1]]]. | In order for plant viruses to enter the host’s genome, it must enter via mechanical transmission. This is because cell walls are too thick for some viruses to penetrate so they rely on transmission through broken cells, animal vectors or through their own seeds[[#References|[16]]]. For Cauliflower Mosaic Virus, Aphids are the most common vector for infection. Aphids possess a needle-like mouthpiece that allows penetration into plants. Aphids then ingest the plants nutrients and sap while also leaving saliva behind in the plant. This saliva can infect the plant if the aphid contains a virus coming from a plant that was ingested beforehand[[#References|[1]]]. | ||
The specific aphid that transports cauliflower mosaic virus is the species Myzus persicae. These fully grown insects grown to be 1.5-2 mm at maturity. They can reproduce at very fast rates and appear during the warmer months to feed on plants. They are polyphageous, which means there is an array of plant species they are able to feed from. Relating to disease, Myzus persicae are known to be able to carry over 100 different types of disease such as cauliflower mosaic virus. Their poisonous saliva can transmit a vast array of diseases from species to species of plant that it chooses to prey on[[#References|[12]]]. | The specific aphid that transports cauliflower mosaic virus is the species Myzus ''persicae'' ('''Fig. 3'''). These fully grown insects grown to be 1.5-2 mm at maturity. They can reproduce at very fast rates and appear during the warmer months to feed on plants. They are polyphageous, which means there is an array of plant species they are able to feed from. Relating to disease, Myzus ''persicae'' are known to be able to carry over 100 different types of disease such as cauliflower mosaic virus. Their poisonous saliva can transmit a vast array of diseases from species to species of plant that it chooses to prey on[[#References|[12]]]. | ||
<br> | <br> | ||
<br> | <br> | ||
[[Image: | [[Image:Cauliflower Mosaic Virus Phylogeny.png|thumb|300px|right|'''Fig 4.''' CaMV Phylogeny [https://www.researchgate.net/figure/Phylogenetic-analysis-of-retrotranscriptase-domains-from-Caulimoviridae-family_fig2_269102694].]] | ||
==Replication== | ==Replication== | ||
The viral dsDNA genome is introduced via an aphid bite. Once inside the host, the viral genome is un-encapsulated and its DNA is transcribed by the host RNA polymerase to make a copy of viral RNA. Host reverse transcriptase then transcribes copies viral RNA back to DNA. Viral RNA also is used to produce gene products such as new capsid proteins and movement proteins in combination with viral DNA to assemble progeny virions. Capsid proteins house genetic material of viruses and offer protection. Movement proteins interact with plasmodesmata in order to allow for transport that otherwise would not occur[[#References|[13]]]. Movement proteins then facilitate movement of progeny virions into uninfected cells of the host plant via plasmodesmata[[#References|[16]]]. | The viral dsDNA genome is introduced via an aphid bite. Once inside the host, the viral genome is un-encapsulated and its DNA is transcribed by the host RNA polymerase to make a copy of viral RNA. Host reverse transcriptase then transcribes copies viral RNA back to DNA. Viral RNA also is used to produce gene products such as new capsid proteins and movement proteins in combination with viral DNA to assemble progeny virions ('''Fig. 4'''). Capsid proteins house genetic material of viruses and offer protection. Movement proteins interact with plasmodesmata in order to allow for transport that otherwise would not occur[[#References|[13]]]. Movement proteins then facilitate movement of progeny virions into uninfected cells of the host plant via plasmodesmata[[#References|[16]]]. | ||
==Immunity and Prevention== | ==Immunity and Prevention== | ||
| Line 52: | Line 52: | ||
<br> | <br> | ||
<br> | <br> | ||
[[Image: | [[Image:CaMV.png|thumb|300px|right|'''Fig. 5''' Domains and their effects on Tobacco and Petunia Species[http://www.biologydiscussion.com/plants/list-of-viruses-found-in-plants-microbiology/65790].]] | ||
==Cauliflower Mosaic Virus in Biotechnology== | ==Cauliflower Mosaic Virus in Biotechnology== | ||
Cauliflower mosaic virus has a very important function in biotechnology. It is because of its efficient promoter that is used to produce cloned genes, that it can be used to create transgenic plants that can take use of this efficiency. That is why now, 10% of cruciferous vegetables have a CaMV infection[[#References|[16]]]. In order to do this, genes can only be inserted in the minor coding regions or regions not necessary for virus production which include open reading frame II and open reading frame VII. If done correctly, the production of progeny viruses will not be affected. A use of this is to insert a dihydroxyl folatereductase gene so that the infected plant will be resistant to methotrexate which otherwise is very toxic to plants[[#References|[15]]]. Producing transgenic plants is done in order to gain benefits such as higher yield, resistance, quality and efficiency which would be hugely impactful to farmers who have been plagued by mosaic viruses and had to throw away a large percentage of their crop[[#References|[11]]]. | Cauliflower mosaic virus has a very important function in biotechnology. It is because of its efficient promoter that is used to produce cloned genes, that it can be used to create transgenic plants that can take use of this efficiency. That is why now, 10% of cruciferous vegetables have a CaMV infection[[#References|[16]]]. In order to do this, genes can only be inserted in the minor coding regions or regions not necessary for virus production which include open reading frame II and open reading frame VII. If done correctly, the production of progeny viruses will not be affected. A use of this is to insert a dihydroxyl folatereductase gene so that the infected plant will be resistant to methotrexate which otherwise is very toxic to plants[[#References|[15]]]. Producing transgenic plants is done in order to gain benefits such as higher yield, resistance, quality and efficiency which would be hugely impactful to farmers who have been plagued by mosaic viruses and had to throw away a large percentage of their crop[[#References|[11]]]. | ||
There are some limitations to using cauliflower mosaic virus in the production of transgenic plants. These limitations include the small margin for error when inserting genes only in open reading frame II and open reading frame VII[[#References|[11]]]. It is also feared that trangenetic plants could induce the evolution of new pararetroviruses similar to how bacteria evolve to be resistance to antibiotics[[#References|[16]]]. | There are some limitations to using cauliflower mosaic virus in the production of transgenic plants. These limitations include the small margin for error when inserting genes only in open reading frame II and open reading frame VII[[#References|[11]]]. It is also feared that trangenetic plants could induce the evolution of new pararetroviruses similar to how bacteria evolve to be resistance to antibiotics[[#References|[16]]]. | ||
Regulatory cis proteins are necessary in interacting which transcription factors to initiate gene expression. The 35S promoter in Cauliflower Mosaic Virus, is used in combination with other 35S subdomains in other plants in order to produce tissue specific gene expression. These other 35S subdomains are different in other plants which means the same combinations of 35S subdomains produce different genes depending on the host plant. The resulting transgenic plant that hosts the Cauliflower Mosaic Virus 35S promoter is expected to differ developmentally in its tissues than the non transgenic plant. Different combinations of 35s subdomains will then be expected to change the development of other plant tissues. Having quick transgenic plant development and easily observed tissue changes, this process is able to be repeated very efficiently and provide more data involving the changes occurring from the CaMV 35s promoter[[#References|[17]]]. | |||
Deletion analysis has been used in order to identify the specific functions of 35S promoter regions. Some regions found in tobacco callus and leaf tissue such as the -343 to -46 region is heavily involved in the activation of transcription. Another region, -90 to +8 was deleted and no effect of leaf tissue occurred unless the -90 to +8 portion is fused with the -343 to -208 regions. This exemplifies how different 35S regions play a role in gene expression which can be used to confer tissue specific gene expression in a multitude of plant species. For example, domain A was found to be involved in root or embryonic root development as well as the meristematic portion of the plant stem and domain B was found to be involved in the upper portions of the plant. These domains are also found in some cases to only be active when they are working synergistically. In tobacco seeds, sub-domain B1 does not change gene expression and change tissue development in seedling growth. However, paired with domain A, differing tissue development and gene expression can be detected. B2 sub-domain paired with the A domain does not affect seed and seedling development at all. Instead, this combination changes in leaf phloem, stem and mature root development. In contrast with the effects of the CaMV promoter in tobacco, petunia plants express genes differently while still using the same 35S promoter. Similarly in tobacco, petunia also requires interaction of the A and B domains in order for many of the B sub-domains to activate gene expression. In contrast, petunia relies on interaction among cis elements unlike tobacco plants. Sub-domain B3 in petunia was also observed to express genes in mature petals without the need of a synergistic relationship with domain A.Without these unique domain relationships, tissue specific gene expression would not be possible[[#References|[17]]]('''Fig. 5'''). | |||
These results could have a huge impact on plant yield, resistance, quality and growth efficiency. The ability to target specific tissues in a multitude of different plants with the cauliflower mosaic virus 35S promoter can facilitate the rise of transgenic plants. Farmers may soon be able to do things such as change leaf area to allow for maximum photosynthetic rate, induce seed germination to increase the length of the growing season or strengthen stems in high wind areas etc. Changing one or all or any other feature of a plant could lead to the eradication of world hunger, reducing poverty in foreign countries, increased food production in response to natural disasters because the possibilities are endless. The 35S promoter provides a mechanism for change that can change the field of biotechnology which it has already contributed too. | |||
<br> | <br> | ||
<br> | <br> | ||
| Line 98: | Line 104: | ||
16. [Slonczewski, Joan L., and John Watkins. Foster. Microbiology: an Evolving Science. 4th ed., W. W. Norton & Company, Inc., 2017.] | 16. [Slonczewski, Joan L., and John Watkins. Foster. Microbiology: an Evolving Science. 4th ed., W. W. Norton & Company, Inc., 2017.] | ||
17. [Benfey, P. N., and N.-H. Chua. “The Cauliflower Mosaic Virus 35S Promoter: Combinatorial Regulation of Transcription in Plants.” Science, vol. 250, no. 4983, 1990, pp. 959–966., doi:10.1126/science.250.4983.959.] | |||
<br><br>Authored for BIOL 238 Microbiology, taught by [mailto:slonczewski@kenyon.edu Joan Slonczewski], 2018, [http://www.kenyon.edu/index.xml Kenyon College]. | <br><br>Authored for BIOL 238 Microbiology, taught by [mailto:slonczewski@kenyon.edu Joan Slonczewski], 2018, [http://www.kenyon.edu/index.xml Kenyon College]. | ||
Latest revision as of 20:26, 11 May 2018
Introduction
By Alan Brennan
The topic of my research is the Cauliflower Mosaic Virus (CaMV). The goal of my research is to identify as well as learn about the structure and function of the Cauliflower Mosaic Virus. Learning of its processes and functions could lead to further studies in transgenic research and increased agricultural efficiency.
Taxonomy
Domain: Virus
- Group: VII
- Family: Caulimoviridae
- Genus: Caulimovirus
- Species: Cauliflower mosaic virus [4]
- Genus: Caulimovirus
- Family: Caulimoviridae
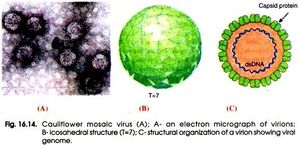
Description and Significance
Mosaic plant diseases are common diseases that occur during warmer months which are caused by plant mosaic viruses. These plant mosaic viruses are identified in plants based on their “mottling” and discoloration of leaves[2]. Viruses may also cause leaf deformity, smaller produce and stunted growth. This could lead to plant death and a reduction of crop production which is why the study of mosaic viruses are essential to improve agriculture by both quantity and quality.
Cauliflower Mosaic Virus (CaMV), specifically, effects the Brassicaceae family and impacts a host range including crops such as broccoli, cabbage, cauliflower and turnips.Cauliflower mosaic virus is classified as a Group VII. pararetrovirus. This means the virus requires transcription from its DNA genome to RNA and then reverse transcriptase to transcribe RNA back into DNA genomes in order to produce progeny virions[16]. It was the first plant virus discovered to utilize a DNA genome and replicate via reverse transcriptase. Cauliflower mosaic virus is usually transmitted from plant to plant via aphid feeding, where virions make their way to insert themselves into the plants nuclear envelope where they inhibit growth and structure of the host.
Cauliflower mosaic virus has been negatively impactful worldwide. Previous studies have shown that CaMV cause a reduction of 25-59% sellable cauliflower heads in Brittany, France as well as a reduction of 60-90% of sellable turnips in the Czech Republic[3]. Breakouts of disease can be devastating to agriculture and national economies. Studying and researching this virus could prevent disease and possibly stop the next potato famine of the cauliflower plant.
Cauliflower Mosaic Virus possesses a highly sought after efficient promoter for gene transcription that can be used in an effort to produce transgenic plants[16]. Transgenic plants are used in order to receive benefits such as producing better yield, quality as well as resistance to insects, diseases and herbicides[11].Currently, about 10% of cruciferous vegetables are infected with Cauliflower Mosaic Virus in order to give the host plant pesticide resistance.
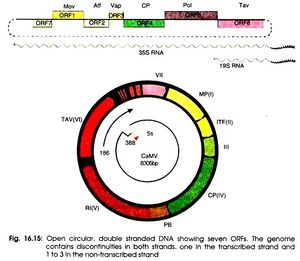
Structure and Genome
Cauliflower Mosaic Virus is classified as an icosahedral (20 sided) virus. This is because of the icosahedral shape of the capsid that surrounds the viral genome. This shape gives the virus 20 symmetrical triangular faces (Fig, 1). This symmetry allows for synthesis efficiency by having the same small number of proteins being produced from a small number genes and from short chromosome sequences. This allows the largest and most complex capsid to be formed from the least amount of resources making it more efficient than a non-symmetrical capsid[16]. The structure of cauliflower mosaic virus is approximately 52 nanometers in diameter and is surrounded by 420 capsid proteins arranged in triangulation T=7[14]. In addition to capsid proteins, caulimoviruses are also surrounded by virus associated proteins[5]. These proteins are responsible for assisting in the binding of the virus to DNA on its c-terminal end[6].
The efficiency of the icosahedral structure allows it so that the genome of CaMV does not even need to encode for more than 7 genes that already have close similarities. Genes differentiate by where their start codons are so that they have different reading frames[16]. The virion that is encapsulated in the capsid structure is made up of around 8,000 base pairs. This double stranded DNA has an open and circular shape (Fig. 2). Also, these strands contain nicks that come from reverse transcription. These nicks are only repaired once these DNA strands enter the host when they form supercoiled molecules responsible for binding to proteins[14].
Cauliflower mosaic virus is well known for its strong constitutive 35S promoter. This promoter runs transcription for the entire cauliflower mosaic virus genome. It is because of its efficiency, it is commonly used for studying transgenic plants in relation to gene transfer vectors. The 600 nucleotide leader sequence of the 35S promoter consists of 8 open reading frames that each possess different functions[15]. There are 6 major coding regions and 2 minor coding regions. Open reading frame I functions to produce movement proteins to assist progeny viruses to pass through plasmodesmata into uninfected cells[7]. Open reading frame II produces aphid transmission factors which is required for the transmission of the virus from the aphid to the plant[8]. Open reading frame III, which previously had no assigned function and is a minor coding region, is believed to promote DNA binding as well as produce structural proteins. ORF IV is responsible for the production of capsid proteins that surround and protect viral genomes. ORF V produces proteins that have proteinases as well as participates in the reverse transcriptase process[8]. Open reading frame VI produces trans activator proteins that promote formation of inclusion bodies. Inclusion bodies are places of viral multiplication needed to produce many progeny virions. The second minor coding region, open reading frame VII, has an unknown function but is speculated to have some function relating to targeting new viruses and bringing them to inclusion bodies (P, Ashwathi.). A very unique function of open reading frame VI that is encoded by the 19S RNA is that it can reinitiate major reading frames on the 35S RNA when this is usually only found to happen in bacteria[15].

Ecology and Pathology
In order for plant viruses to enter the host’s genome, it must enter via mechanical transmission. This is because cell walls are too thick for some viruses to penetrate so they rely on transmission through broken cells, animal vectors or through their own seeds[16]. For Cauliflower Mosaic Virus, Aphids are the most common vector for infection. Aphids possess a needle-like mouthpiece that allows penetration into plants. Aphids then ingest the plants nutrients and sap while also leaving saliva behind in the plant. This saliva can infect the plant if the aphid contains a virus coming from a plant that was ingested beforehand[1].
The specific aphid that transports cauliflower mosaic virus is the species Myzus persicae (Fig. 3). These fully grown insects grown to be 1.5-2 mm at maturity. They can reproduce at very fast rates and appear during the warmer months to feed on plants. They are polyphageous, which means there is an array of plant species they are able to feed from. Relating to disease, Myzus persicae are known to be able to carry over 100 different types of disease such as cauliflower mosaic virus. Their poisonous saliva can transmit a vast array of diseases from species to species of plant that it chooses to prey on[12].
Replication
The viral dsDNA genome is introduced via an aphid bite. Once inside the host, the viral genome is un-encapsulated and its DNA is transcribed by the host RNA polymerase to make a copy of viral RNA. Host reverse transcriptase then transcribes copies viral RNA back to DNA. Viral RNA also is used to produce gene products such as new capsid proteins and movement proteins in combination with viral DNA to assemble progeny virions (Fig. 4). Capsid proteins house genetic material of viruses and offer protection. Movement proteins interact with plasmodesmata in order to allow for transport that otherwise would not occur[13]. Movement proteins then facilitate movement of progeny virions into uninfected cells of the host plant via plasmodesmata[16].
Immunity and Prevention
In order to defend against viral infection of cauliflower mosaic virus, plants must depend on their natural defenses or risk death. In a situation using cauliflower as an example, in farms having cauliflower as a cash crop, the risk of viral infection is very high. It is because they are all grown in proximity and in the same conditions where random mutation is not favored, it would not be favorable to viral resistance. In this scenario, breeding with mutated cauliflower would be the best chance of gaining resistance[16].
Plants also must rely on their own immune systems. If a plant can utilize its ability to interfere with gene expression then virions will stop being replicated inside of the host. The plant is able to recognize foreign mRNA and stop gene expression before the plant continues producing for the virus[16].
There is no cure for cauliflower mosaic virus. Once a plant is infected, then it is too late. Prevention is the only way to prevent agricultural or gardening losses. Since cauliflower mosaic virus is usually spread by aphids, so any sort of pest control can be beneficial in reducing infection. Netting or pest control products may keep virus carrying insects at bay. Since this virus can be transported through any opening in plants such as abrasions or cuts, disinfecting tools, equipment and anything that contacts plants will reduce infections. Some things such as damp conditions may also contribute to faster/easier transmission of cauliflower mosaic virus. It is best to get rid of infected plants immediately to reduce exposure to plants nearby. Also, get rid of seeds coming from infected plants because CaMV could be transmitted to plant offspring. Keeping weeds away from the farm or garden can also eliminate virus harboring organisms. Finally, there are varieties of plants with resistance to caulimoviruses that are unable to get infected in the first place[9][10].

Cauliflower Mosaic Virus in Biotechnology
Cauliflower mosaic virus has a very important function in biotechnology. It is because of its efficient promoter that is used to produce cloned genes, that it can be used to create transgenic plants that can take use of this efficiency. That is why now, 10% of cruciferous vegetables have a CaMV infection[16]. In order to do this, genes can only be inserted in the minor coding regions or regions not necessary for virus production which include open reading frame II and open reading frame VII. If done correctly, the production of progeny viruses will not be affected. A use of this is to insert a dihydroxyl folatereductase gene so that the infected plant will be resistant to methotrexate which otherwise is very toxic to plants[15]. Producing transgenic plants is done in order to gain benefits such as higher yield, resistance, quality and efficiency which would be hugely impactful to farmers who have been plagued by mosaic viruses and had to throw away a large percentage of their crop[11]. There are some limitations to using cauliflower mosaic virus in the production of transgenic plants. These limitations include the small margin for error when inserting genes only in open reading frame II and open reading frame VII[11]. It is also feared that trangenetic plants could induce the evolution of new pararetroviruses similar to how bacteria evolve to be resistance to antibiotics[16].
Regulatory cis proteins are necessary in interacting which transcription factors to initiate gene expression. The 35S promoter in Cauliflower Mosaic Virus, is used in combination with other 35S subdomains in other plants in order to produce tissue specific gene expression. These other 35S subdomains are different in other plants which means the same combinations of 35S subdomains produce different genes depending on the host plant. The resulting transgenic plant that hosts the Cauliflower Mosaic Virus 35S promoter is expected to differ developmentally in its tissues than the non transgenic plant. Different combinations of 35s subdomains will then be expected to change the development of other plant tissues. Having quick transgenic plant development and easily observed tissue changes, this process is able to be repeated very efficiently and provide more data involving the changes occurring from the CaMV 35s promoter[17].
Deletion analysis has been used in order to identify the specific functions of 35S promoter regions. Some regions found in tobacco callus and leaf tissue such as the -343 to -46 region is heavily involved in the activation of transcription. Another region, -90 to +8 was deleted and no effect of leaf tissue occurred unless the -90 to +8 portion is fused with the -343 to -208 regions. This exemplifies how different 35S regions play a role in gene expression which can be used to confer tissue specific gene expression in a multitude of plant species. For example, domain A was found to be involved in root or embryonic root development as well as the meristematic portion of the plant stem and domain B was found to be involved in the upper portions of the plant. These domains are also found in some cases to only be active when they are working synergistically. In tobacco seeds, sub-domain B1 does not change gene expression and change tissue development in seedling growth. However, paired with domain A, differing tissue development and gene expression can be detected. B2 sub-domain paired with the A domain does not affect seed and seedling development at all. Instead, this combination changes in leaf phloem, stem and mature root development. In contrast with the effects of the CaMV promoter in tobacco, petunia plants express genes differently while still using the same 35S promoter. Similarly in tobacco, petunia also requires interaction of the A and B domains in order for many of the B sub-domains to activate gene expression. In contrast, petunia relies on interaction among cis elements unlike tobacco plants. Sub-domain B3 in petunia was also observed to express genes in mature petals without the need of a synergistic relationship with domain A.Without these unique domain relationships, tissue specific gene expression would not be possible[17](Fig. 5).
These results could have a huge impact on plant yield, resistance, quality and growth efficiency. The ability to target specific tissues in a multitude of different plants with the cauliflower mosaic virus 35S promoter can facilitate the rise of transgenic plants. Farmers may soon be able to do things such as change leaf area to allow for maximum photosynthetic rate, induce seed germination to increase the length of the growing season or strengthen stems in high wind areas etc. Changing one or all or any other feature of a plant could lead to the eradication of world hunger, reducing poverty in foreign countries, increased food production in response to natural disasters because the possibilities are endless. The 35S promoter provides a mechanism for change that can change the field of biotechnology which it has already contributed too.
Conclusion
Cauliflower mosaic virus is one of many mosaic viruses that infect crops and plants across the globe. Mosaic viruses have many forms such as tobacco mosaic virus and cucumber mosaic virus to name a few. With cauliflower mosaic virus having a uniquely efficient promoter, it allows researchers to use it to make other plant aspects efficient by the use of an administered viral infection. Processes such as RNA interference could be better understood and utilized to fight off cancer cells or other diseases in humans[16]. The knowledge of the cauliflower mosaic virus and its promoter could instigate research that could help feed countries during famine. It could help keep the world’s population fed by keeping up with our demand for resources by making plant growth more efficient. It could also improve lives of farmers by not having them see drooping and mottled leaves when it is time to harvest and wasting time, energy, space and food that could otherwise be used.
References
1.[“Aphid-Transmitted Viruses in Vegetable Crops: Integrated Virus Disease Management.” Pest and Disease Management | Fact Sheets | Resources | Soil Wealth Integrated Crop Protection, Soil Wealth Integrated Crop Protection]
2. [Britannica, The Editors of Encyclopaedia. “Mosaic.” Encyclopædia Britannica, Encyclopædia Britannica, Inc., 15 Dec. 2017, www.britannica.com/science/mosaic-plant-disease. ]
3. [“Cauliflower Mosaic (Cauliflower Mosaic Virus).” Cauliflower Mosaic (Cauliflower Mosaic Virus), Plantwise.org, www.plantwise.org/KnowledgeBank/Datasheet.aspx?dsid=15099.]
4. [“Cauliflower Mosaic Virus.” Cauliflower Mosaic Virus (Cauliflower Mosaic), Cabi.org, www.cabi.org/isc/datasheet/15099.]
5. [“Caulimovirus.” Viralzone, viralzone.expasy.org/119?outline=all_by_species. ]
6. [“InterPro.” Caulimovirus Virion-Associated Protein (IPR004986) < InterPro < EMBL-EBI, InterPro, www.ebi.ac.uk/interpro/entry/IPR004986.]
7. [Kobayashi, Kappei. “Kappei Kobayashi.” Journal of Virology, 1 Aug. 2003, jvi.asm.org/content/77/15/8577.full.]
8. [Leh, V. “Aphid Transmission of Cauliflower Mosaic Virus Requires the Viral PIII Protein.” The EMBO Journal, vol. 18, no. 24, 1999, pp. 7077–7085., doi:10.1093/emboj/18.24.7077.]
9. [“Mosaic Viruses.” Old Farmer's Almanac, Old Farmer’s Almanac, www.almanac.com/pest/mosaic-viruses.]
10. [“Mosaic Virus: Symptoms, Treatment and Control.” Planet Natural, Planet Natural, 2017, www.planetnatural.com/pest-problem-solver/plant-disease/mosaic-virus/.]
11. [Mondal, Sudhadip. “8 Main Advantages of Transgenic Plant | Genetics.” Biology Discussion, Biology Discussion, 12 July 2016, www.biologydiscussion.com/plants/transgenic-plants/8-main-advantages-of-transgenic-plant-genetics/38097.]
12. [“Myzus Persicae.” Myzus-Persicae - Bayer - Crop Science, Bayer, www.cropscience.bayer.com/en/crop-compendium/pests-diseases-weeds/pests/myzus-persicae.]
13. [Niehl, Annette, and Manfred Heinlein. “Cellular Pathways for Viral Transport through Plasmodesmata.” SpringerLink, Springer Vienna, 2 Dec. 2010, link.springer.com/article/10.1007%2Fs00709-010-0246-1#Sec9.]
14. [P, Ashwathi. “List of Viruses Found in Plants | Microbiology.” Biology Discussion, Biology Discussion, 28 Nov. 2016, www.biologydiscussion.com/plants/list-of-viruses-found-in-plants-microbiology/65790.]
15. [Padmaratinam Follow. “Cauliflower Mosaic Virus.” LinkedIn SlideShare, LinkedIn, 12 Mar. 2016, www.slideshare.net/Padmaratinam/cauliflower-mosaic-virus.]
16. [Slonczewski, Joan L., and John Watkins. Foster. Microbiology: an Evolving Science. 4th ed., W. W. Norton & Company, Inc., 2017.]
17. [Benfey, P. N., and N.-H. Chua. “The Cauliflower Mosaic Virus 35S Promoter: Combinatorial Regulation of Transcription in Plants.” Science, vol. 250, no. 4983, 1990, pp. 959–966., doi:10.1126/science.250.4983.959.]
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2018, Kenyon College.
