Comamonas badia: Difference between revisions
No edit summary |
No edit summary |
||
| Line 28: | Line 28: | ||
[[Image:Phylogenic_Tree_of_Comamonas_badia.jpeg|thumb|300px|left|Figure 3. Simplified phylogenetic tree made comparing 16S rDNA sequence of ''Comamonas badia'' to some members of the β-proteobacteria. ''Alcaligenes faecalis'' is used as an outgroup. Source: Image is derived from Fig. 2 from Tago Y. and Yokota A. (2004) [[#References|[1]]]] | [[Image:Phylogenic_Tree_of_Comamonas_badia.jpeg|thumb|300px|left|Figure 3. Simplified phylogenetic tree made comparing 16S rDNA sequence of ''Comamonas badia'' to some members of the β-proteobacteria. ''Alcaligenes faecalis'' is used as an outgroup. Source: Image is derived from Fig. 2 from Tago Y. and Yokota A. (2004) [[#References|[1]]]] | ||
It was only recently in 2004 that C. badia was introduced and recognized as a novel species, using comparative analysis of almost complete 16S rDNA gene sequence and G+C content of DNA on the strain IAM 14839<sup>T</sup>. The complete genome of ''C. badia'' has yet to be sequenced [[#References|[1]]][[#References|[9]]]. | It was only recently in 2004 that C. badia was introduced and recognized as a novel species, using comparative analysis of almost complete [http://en.wikipedia.org/wiki/16S_rDNA 16S rDNA] gene sequence and [http://en.wikipedia.org/wiki/GC-content G+C content] of DNA on the strain IAM 14839<sup>T</sup>. The complete genome of ''C. badia'' has yet to be sequenced [[#References|[1]]][[#References|[9]]]. | ||
=References= | =References= | ||
Revision as of 03:08, 13 December 2012
Classification
Higher Order Taxa
Bacteria (Kingdom), Proteobacteria (Phylum), Betaproteobacteria (Class), Burkholderiales (Order), Comamonadaceae (Family), Comamonas(Genus)
Species
Comamonas badia
Description
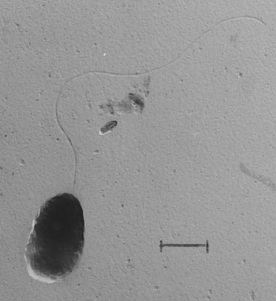
Comamonas Badia is a gram negative, rod-shaped, strictly aerobic, and highly motile bacteria with a polar flagellum and was isolated from phenol-adapted activated sludge from a factory sludge from Japan [1]. It was originally classified under the genus Pseudomonas [3][4]. Based on 16S rRNA gene sequence and G+C content, C. badia was determined to represent its own genus, Comamonas. This novel one was first represented by the species C. badia, type-strain IAM 14839T [1][9].
Comamonas badi forms pink colonies when cultured in Luria slant media and forms visible flocs in Luria broth media, unlike other Comamonas species which do not form flocs [2]. Comamonas badia grows at a very slow rate in a flocculated state with a generation time of about 12 hours at optimal temperatures (27-30°C) in LB media and produces few free-floating cells. When cultured in broths, the supernatant is usually clear [4]. This bacterium grows the slowest at 20°C (19 hours for generation time) and does not grow at 37°C (average human body temperature), unlike other Comamonas species which do undergo growth at 37°C [1]. For floc formation, C. badia grows embedded in a film mesh structure made out of mucopolysaccharides which consists of glucosamine, glucose, mannose, galactose and rhamnose. Only about 10% of the total polysaccharide content found with C. badia is involved with floc formation [3]. The ability to form flocs gives this species a protection advantage from predators compared to free roaming cells, allows other bacteria to form on the flocs, and prevents the organism from being washed out of the system that it is in, i.e. activated sludge [10].
Significance

It has been found that C. badia has moderate phenol tolerance and moderate phenol degradation capabilities. C. badia existing in a floc-formation is also the most predominating bacteria species in activated sludge and is involved indegrading highly toxic and carcinogenic phenol compounds in coke wastewater [2]. Being able to tolerate and degrade such toxic compounds, the ability to form flocs is an integral part this microbe in activated sludge to treat wastewaters [10]. C. badia has a unique ability in which it is responsible for its own deflocculation by producing an exoenzyme. Various commercial enzymes are unable to induce deflocculation in C. badia and therefore the bacteria’s life activities are determined by their own accord via polysaccharide-synthesizing enzymes and deflocculating enzymes [5].
Phylogenetics
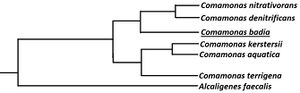
It was only recently in 2004 that C. badia was introduced and recognized as a novel species, using comparative analysis of almost complete 16S rDNA gene sequence and G+C content of DNA on the strain IAM 14839T. The complete genome of C. badia has yet to be sequenced [1][9].
References
(1) Tago Y., Yokota A. “Comamonas badia sp nov., a Floc-forming Bacterium Isolated from Activated Sludge.” Journal of General and Applied Microbiology, 2004, DOI: 10.2323/jgam.50.243
(2) Felföldi T., Székely A., Gorál R., Barkács K., Scheirich G., András J, Rácz A., Márialigeti K. “Polyphasic Bacterial Community Analysis of an Aerobic Activated Sludge Removing Phenols and Thiocyanate from Coke Plant Effluent.” Bioresource Technology, 2010, DOI: 10.1016/j.biortech.2009.12.053
(3) Tago Y., Aida K. “Exocellular Mucopolysaccharide Closely Related to Bacterial Floc Formation” Applied and Environmental Microbiology, 1977, ISSN: 0099-2240
(4) Tago Y., Kuraishi H., Aida K. “The Formation of a Model Floc able to Decompose Phenol by the Mixed Culture of Bacteria Isolated from Activated Sludge.” Journal of General and Applied Microbiology, 1975, DOI: 10.2323/jgam.21.41
(5) Tago Y., Aida K. “The Deflocculating Enzyme Produced by a Floc-Forming Bacterium.” Journal of General and Applied Microbiology, 1975, DOI: 10.2323/jgam.21.365
(6) Blöthe M., Roden E. “Composition and Activity of an Autotrophic Fe(II)-Oxidizing, Nitrate-Reducing Enrichment Culture” Applied and Environmental Microbiology, 2009, DOI: 10.1128/AEM.01742-09
(7) Mahendran B., Lishman L., Liss S. “Structural, Physicochemical and Microbial Properties of Flocs and Biofilms in Integrated Fixed-Film Activated Sludge (IFFAS) Systems” Water Research (Oxford), 2012, DOI: 10.1016/j.watres.2012.05.058
(8) Zita A., Hermansson M. “Effects of Bacterial Cell Surface Structures and Hydrophobicity on Attachment of Activated Sludge Flocs” Applied and Environmental Microbiology, 1997, ISSN: 0099-2240
(9) Stackebrandt E., Goebel B. “Taxonomic Note: A place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology” International Journal of Systematic Bacteriology, 1994, ISSN: 0020-7713
(10) Raszka A., Chorvatova M., Wanner J. “The Role and Significance of Extracellular Polymers in Activated Sludge. Part I: Literature Review” Acta Hydrochimica et Hydrobiologica (Weinheim an der Bergstrasse, Germany), 2006, DOI: 10.1002/aheh.200500640
