Finegoldia magna: Difference between revisions
No edit summary |
|||
| Line 82: | Line 82: | ||
=='''Genome Structure'''== | =='''Genome Structure'''== | ||
''Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence?'' | ''Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence?'' | ||
Finegoldia magna has been sequenced and specifically, strain ATCC29328 has been adequately classified. The genome consists of a single, circular, 1.8Mb chromosome along with a 1.9Mb megaplasmid (pPEP1). Colonies of F. magna appear pyramidal in the center, opaque, shiny, whitish-yellow, and gray on the outer ring of the spherical colonies. Compared to other GPACs, the Guanine-Cytosine nucleotide content present within the genome is relatively small at 32.3% and 29.7% with respect to the chromosome and plasmid (pPEP1). The genome is also comprised of unusually high aminopeptidase activity as well as many sortase genes contributing to the microbes pathogenicity; 7 sortases within the plasmid and 4 on the chromosome. | Finegoldia magna has been sequenced and specifically, strain ATCC29328 has been adequately classified. The genome consists of a single, circular, 1.8Mb chromosome along with a 1.9Mb megaplasmid (pPEP1). Colonies of F. magna appear pyramidal in the center, opaque, shiny, whitish-yellow, and gray on the outer ring of the spherical colonies. Compared to other GPACs, the Guanine-Cytosine nucleotide content present within the genome is relatively small at 32.3% and 29.7% with respect to the chromosome and plasmid (pPEP1). The genome is also comprised of unusually high aminopeptidase activity as well as many sortase genes contributing to the microbes pathogenicity; 7 sortases within the plasmid and 4 on the chromosome. | ||
Revision as of 17:00, 29 April 2022
Classification

Domain: Bacteria
Phylum: Firmicutes
Class: Clostridia
Order: Eubacteriales
Family: Peptoniphilaceae
Genus: Finegoldia
Species
Finegoldia magna
F. magna was previously classified as Peptostreptococcus anaerobius, and it was recognized as a unique species as recently as 1998. [4]
|
NCBI: Taxonomy [5] |
Description and Significance
Describe the appearance, habitat, etc. of the organism, and why you think it is important.
"Finegoldia magna" is a slow-growing, non-spore-forming, Gram-positive anaerobic coccus (GPAC) bacteria.
- F. magna is a major part of healthy skin and mucosal surface microflora. It has been shown to be the most commonly-isolated anaerobe from skin specimens [6] , and it is prevalent in chronic wounds, such as diabetic and pressure ulcers. [7] It is also commonly found as the culprit of secondary infections following prosthetic joint replacement or cardiac valve replacement surgeries. [8] ] The importance of F. magna as a human pathogen is insufficiently recognized, especially as it has been recently implicated in a case of toxic shock syndrome. [9]
- The color of the colonies is usually translucent, but it can appear whitish to yellow or gray. [10]
- The Genus namesake is in remembrance of the American microbiologist Sydney Martin Finegold.
Genome Structure
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence?
Finegoldia magna has been sequenced and specifically, strain ATCC29328 has been adequately classified. The genome consists of a single, circular, 1.8Mb chromosome along with a 1.9Mb megaplasmid (pPEP1). Colonies of F. magna appear pyramidal in the center, opaque, shiny, whitish-yellow, and gray on the outer ring of the spherical colonies. Compared to other GPACs, the Guanine-Cytosine nucleotide content present within the genome is relatively small at 32.3% and 29.7% with respect to the chromosome and plasmid (pPEP1). The genome is also comprised of unusually high aminopeptidase activity as well as many sortase genes contributing to the microbes pathogenicity; 7 sortases within the plasmid and 4 on the chromosome.
The isolation of F. magna ATCC29328 was more easily identified and classified than other strains of F. magna due to the presence of increased aminopeptidase activity and the outstanding number of sortase genes present within the plasmid. These genomic characteristics give light to the bacteriums’ enhanced interaction with hosts and its ability to uptake amino acids as energy sources.
Cell Structure, Metabolism and Life Cycle
Interesting features of cell structure; how it gains energy; what important molecules it produces.
Cell Structure
- Finegoldia magna is a Gram-positive anaerobic coccus (GPAC) bacteria, in which "coccus" indicates a spherical cell shape. Gram-positive indicates a bacterium with thick peptidoglycan cell walls.
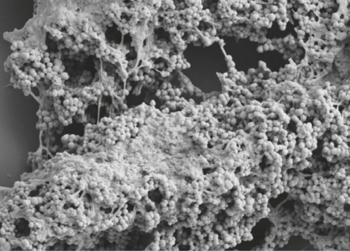
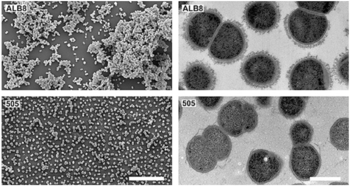
- It has a cell size that varies from 0.8 µm and 1.6 µm in diameter, and cells occur predominantly in masses or clusters but occasionally in short chains or pairs, as in Figure 4. [3]
- This bacteria has sortase-dependent pilli, which are hair-like structures important for bacterial adhesion and colonization (i.e., important for biofilm formation) as well as host infection. [11] Sortases are enzymes that modify surface proteins of the cell wall.
Metabolism
- As an anaerobe, F. magna requires an oxygen-free environment to grow. However, in a 1998 experiment by Murdoch et al., F. magna isolates were plated on agar and exposed to air. After 48 hours, some clusters survived, indicating some level of oxygen tolerance. [12]
- F. magna produces acetic acid as a major fermentation product. It produces weak acid utilizing fructose, while a few strains use glucose as a carbon source. Peptones and amino acids are also used an energy sources. It can produce ammonia from glycine, threonine, and serine. [13]
- Aminopeptidases (i.e. catalyze cleavage of amino acids) have been reported but are not well-studied. [14]
Survival and Life Cycle
- F. magna is susceptible to antibiotics used to treat anaerobic infections. However, it shows some resistance (10-20%) to clindamycin, metronidazole, penicillin, while even higher resistance (>20%) to erythromycin and tetracycline. Its ability to form mono-species and dual-species biofilms may lend greater protection against host immune defenses and antibiotics. [1]
- The nutritional requirements of cultured GPAC organisms, including F. magna are understudied. Commercial media tend to vary in their ability to support growth. However, Fastidious Anaerobe Agar has consistently proved most successful. Sodium oleate, vitamin K, and hemin supplements are not necessary. F. magna can live at room temperature for months to years on adequate media. [12]
- Cocci bacteria, including F. magna, replicate asexually through binary fission and can divide from different planes to form a variety of arrangements (as compared to bacilli, which divide from the horizontal axis).
Ecology and Pathogenesis
- Forms a commensalism relation with host as it is abundant on the skin surface with no benefit to the host, however relationship can quickly turn pathogenic as F. magna will colonize any non-sterile body surface leading to infection when open wounds are present
Virulence Factors
- Protein L: Binds to neutrophils and prevents attack
- Pab: Binding of HSA, causes increased growth rate and cell density
- FAF: Binds to BM-40 increases adherence of cells and aggregation
- SufA: Degradation of fibrinogen
Infections
- F. magna colonizes any non-sterile body surface
- Found in prosthetic joint infections but can appear in septic arthritis and other infections as well
- Infections can be divided into two clinical entities[8]
- First entity is characterized by post traumatic infection with open fractures. Generally a polymicrobial infection with F. magna being found with other anaerobic species
- The second clinical entity is typically a nosocomial infection and involves patients who have received a hip or knee prosthesis followed by pain, mild fever, inflammation, and the eventual loosening of the prosthesis at the bone interface. In these cases, F. magna was the sole etiological agent
- Symptoms of infection can start anywhere anywhere between 4-12 months for prosthetics, but <4 for other kinds
Biofilms
- Finegoldia Magna forms a slow -growing biofilm in the oral cavity and in the intestines of the immunocompromised and on prosthetics/infection[1]
- Color of the biofilm formed is often translucent but can also be white or gray
- The biofilm formed is strongly adherent and is suspected to be a source of protection from host immune defenses
- Can form dual-species biofilm with both B. fragilis and C. difficile
References
Authors
Page authored by Susan Reed, Nitesh Naren, Matt Millikin, students of Prof. Jay Lennon at Indiana University.

