Halorhabdus utahensis
Classification
Archaea; Euryarchaeota; Halobacteria; Halobacteriales; Halobacteriaceae; Halorhabdus
Species
Halorhabdus utahensis
Description and Significance
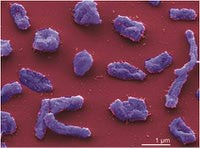
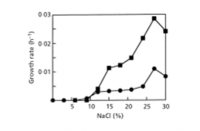
Halorhabdus (salt-loving rod) utahensis (pertaining to the state of Utah) was isolated from a sediment sample that was collected in the southern arm of the hypersaline Great Salt Lake, Utah, USA. It is a motile, gram-negative, extreamly halophilic archaeon that forms red, circular colonies. It grows in temperatures between 17 and 55°C, with optimal growth occurring at 50°C. It can also grow over a pH range of 5.5-8.5 with the optimal pH value between 6.7 and 7.1. Halorhabdus utahensis is of interest due to its ability to grow over a wide range of salinity. It can grow anywhere from 6-30% NaCl with an optimum of an astounding 27% (2).
The cells of Halorhabdus utahensis are extremely pleomorphic, exhibiting anything from irregular coccoid or ellipsoid to triangular, club-shaped or rod-shaped forms. The rod-shaped and ellipsoid cells are 2-10 * 0.5-1 µm and 1-2 * 1 µm in size, respectively, and the spherical cells have a diameter of approximately 1 µm. The archaeon utilizes only a limited range of substrates, such as glucose, xylose, and fructose, for growth, and is unique in its inability to utilize yeast extract or peptone. Other substances that do not stimulate the organism's growth include organic acids, amino acids, alcohols, glycogen, and starch (2).
A recent research (2011) paper described the significance of Halorhabdus utahensis as ionic liquid(IL)-resistant cellulase. This is due to their adaption to its extreme environment. Halorhabdus utahensis enzymes are hypothesized to be useful in the identification of IL-resistant cellulases, having a highly enriched, negatively charged acidic amino acids. These amino acids on the surface are hypothesized to form a shell that stabilizes the enzyme through contact with salt ions, which is found in Halorhabdus utahensis environment (3).
Genome Structure
The entire genome of Halorhabdus utahensis has been sequenced by the DOE Joint Genome Institute (JGI) and is accessible in Genbank. It was sequenced using a combination of Sanger sequencing and also 454 where the error rate of the completed genome sequence is less than 1/100,000. It has been determined that Halorhabdus utahensis contains one main circular chromosome that is 3,116,795 bp long and is comprised of 62.90% G+C content. There are approximately 3,075 genes of which 3,027 are protein encoding genes, 48 are RNA genes, 29 are pseudogenes, and one is an rRNA operon. Some of the genes help to encode Halorhabdus utahensis ability to become pleomorphic (multiple structural forms). It does not need any amino acids to grow and therefore is prototrophic for their acquisition (1). This characteristic utilizes several genes along with the cells resistance to ampicillin, carbenicillin, chloramphenicol, erythromycin, gentamicin, kanamycin, nalidixic acid, neomycin, penicillin, polymyxins, rifampicin, streptomycin, and tetracycline (2).
Cell Structure, Metabolism and Life Cycle
Halorhabdus utahensis is pleomorphic, which means its cells can take on multiple shapes. It is usually in a rod shaped form, particularly in its younger years and later in life it become increasingly pleomorphic (2). Due to its pleomorphic nature it can appear as ellipsoids and spheres, ranging from 1-2X1 micro meters. The organisms are mobile and have a flagellum, ranging in size from 3 to 5 micro meters (1). The cells can be distinguished because of their gram-negative trait. Furthermore, Halorhabdus utahensis has a red color and the pigments are not precisely known but presumed to be, “carotenoids, probably bacterioruberins” (1). The outer cell layers are proteinaceous (1).
Growth of the cell does not require amino acids, but does require nitrogen and a carbon source (2). The metabolism of Halorhabdus utahensis is aerobic, but is also a facilitative anaerobe. It receives its energy from carbohydrates where the carbon source is from fructose, xylose, and glucose. Not important to the growth of the cell, however a side product of it metabolism is the reduction of nitrate to nitrite (1). It has been found that it may grow anaerobically by fermentation of glucose (1).
Ecology and Pathogenesis
Halorhabdus utahensis was isolated from sediment found in the Great Salt Lake in Utah, USA (2). This microbe is extremely halophilic and is found in an environment with high levels of NaCl. When grown in an anaerobic environment with glucose nitrate was reduced to nitrite, although this did not stimulate the microbe’s growth (2). There were no signs of Halorhabdus utahensis engaging in any kinds of symbiosis with other forms of life.
Halorhabdus utahansis has no pathogenicity towards other organisms (1). However it was found that Halorhabdus utahensis was susceptible to bacitracin and novobiocin but resistance to ampicillin, carbenicillen, chloramphenicol, erythromycin, gentamicin, kanamycin, calidixic acid, neomycin, penicillin, polymyxins, rifampicin, streptomycin, and tetracycline (2).
References
1. Anderson, I., Tindall, B., Pomrenke, H., & Goker, M. (2009). Complete genome sequence of Halorhabdus utahensis type strain (AX-2T). Standards in Genomic Sciences, (1), 218-225.
2. Waino, M., Tindall, B., & Ingvorsen, K. (2000). Halorhabdus utahensis gen. nov., sp. nov., an aerobic, extremely halophilic member of the archaea from Great Salt Lake, Utah. International Journal of Systematic Evolutionary Microbiology, (50), 183-190.
3. Zhang, Tao, Supratim Datta, Jerry Eichler, and Natalia Ivanova. “Identification of a Haloalkaliphilic and Thermostable Cellulase with Improved Ionic Liquid Tolerance.” Green Chemistry 13 (2011): 2083-2090.
Author
Page authored by Elliot Swift, Mark Sherrill, Matt Stoloff, Hannah Arnett, student of Prof. Jay Lennon at Michigan State University.
<-- Do not remove this line-->