Human Chromosomal Integration of Latent State Human Herpes Virus 6 (HHV-6): Difference between revisions
| Line 7: | Line 7: | ||
==Genomic Structure of HHV-6== | ==Genomic Structure of HHV-6== | ||
[[Image:HHV-6 Genome.jpeg|thumb|300px|left| [http://cmr.asm.org/cgi/content/full/18/1/217/F1 Figure 2.] Genomic organization of HHV-6B. The asterisk indicates the start of lytic genomic replication. Viral telomeric sequences are indicated by green bars T1 and T2. Open arrows represent protein coding regions.]] | [[Image:HHV-6 Genome.jpeg|thumb|300px|left| [http://cmr.asm.org/cgi/content/full/18/1/217/F1 Figure 2.] Genomic organization of HHV-6B. The asterisk indicates the start of lytic genomic replication. Viral telomeric sequences are indicated by green bars T1 and T2. Open arrows represent protein coding regions.]] | ||
<br> | |||
HHV-6 is an enveloped, linear double stranded DNA virus. The genome is 160 to 162 kB in size, with a 143-144 kB unique region (1, 2). The unique region is flanked by 8-9 kB terminal direct repeats at each end, which contain repeats of the hexanucleotide sequence GGGTTA, identical to the telomeric sequences of vertebrate chromosomes (3, 4). | |||
<br> | |||
<br> | |||
The genome has a cluster of seven gene blocks that are conserved among all herpesviruses, as well as the US22 gene family, conserved in other β-herpesviruses such as CMV and HHV-7 (5, 6). Although the function of the US22 gene family is not fully understood, it has been found that members of this gene family may act as transactivators, enhancing and increasing HHV-6 viral gene expression (7). | |||
<br> | |||
<br> | |||
The products of HHV-6 genes U27, U41, U43, U74, U77, and U73 are involved in viral replication. The protein product of gene U27 encodes the DNA polymerase processivity factor, binding to DNA polymerase and thus, enhances the rate of viral genome transcription. Gene U41 encodes a single stranded DNA binding protein, also facilitating DNA polymerase's interaction with the viral genome. The protein products of genes U43, U74, and U77 produce a viral helicase-primase complex that facilitates replication of the viral genome. Additionally, the origin-binding protein produced by gene U73 also aids in genomic DNA replication. HHV-6 attachment to host cells is facilitated by viral surface proteins gB and gH, encoded by viral genes U39 and U48, respectively (8). | |||
<br> | |||
<br> | |||
Unique to HHV-6 among all other herpesviruses are genes U83 and U94. Gene U83 encodes a chemokine that uses signaling mechanisms such as calcium fluxes to recruit lymphocytes and macrophages for productive or latent infection (9). Gene U94 encodes protein product RepH6, which binds the human TATA-binding protein, where it has an effect on regulation of viral gene expression and viral DNA replication. Furthermore, RepH6 has been shown to play a role in latent stage infections, as it is transcribed in latently infected lymphocytes (10). | |||
<br> | |||
<br> | |||
There are two variants of HHV-6; HHV-6A and HHV-6B. HHV-6A and HHV6B share an overall nucleotide sequence identity of approximately 90%. HHV-6A has 110 ORFs, while HHV-6B has 119 ORFs. Major genomic differences between the two strains lie in the direct repeat regions near the ends of the viral genome, as well as in the proteins encoded by genes U86-U100 in the early stage of infection. The differences observed in early stage viral proteins are the result of the splicing patterns unique to each viral variant (11, 12). | |||
==HHV-6 Replication Cycle== | ==HHV-6 Replication Cycle== | ||
Revision as of 04:14, 2 November 2010
By: Kerri-Lynn Conrad
Introduction
Genomic Structure of HHV-6
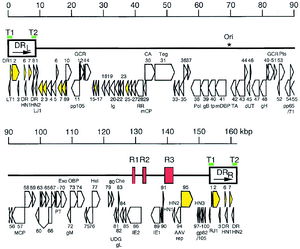
HHV-6 is an enveloped, linear double stranded DNA virus. The genome is 160 to 162 kB in size, with a 143-144 kB unique region (1, 2). The unique region is flanked by 8-9 kB terminal direct repeats at each end, which contain repeats of the hexanucleotide sequence GGGTTA, identical to the telomeric sequences of vertebrate chromosomes (3, 4).
The genome has a cluster of seven gene blocks that are conserved among all herpesviruses, as well as the US22 gene family, conserved in other β-herpesviruses such as CMV and HHV-7 (5, 6). Although the function of the US22 gene family is not fully understood, it has been found that members of this gene family may act as transactivators, enhancing and increasing HHV-6 viral gene expression (7).
The products of HHV-6 genes U27, U41, U43, U74, U77, and U73 are involved in viral replication. The protein product of gene U27 encodes the DNA polymerase processivity factor, binding to DNA polymerase and thus, enhances the rate of viral genome transcription. Gene U41 encodes a single stranded DNA binding protein, also facilitating DNA polymerase's interaction with the viral genome. The protein products of genes U43, U74, and U77 produce a viral helicase-primase complex that facilitates replication of the viral genome. Additionally, the origin-binding protein produced by gene U73 also aids in genomic DNA replication. HHV-6 attachment to host cells is facilitated by viral surface proteins gB and gH, encoded by viral genes U39 and U48, respectively (8).
Unique to HHV-6 among all other herpesviruses are genes U83 and U94. Gene U83 encodes a chemokine that uses signaling mechanisms such as calcium fluxes to recruit lymphocytes and macrophages for productive or latent infection (9). Gene U94 encodes protein product RepH6, which binds the human TATA-binding protein, where it has an effect on regulation of viral gene expression and viral DNA replication. Furthermore, RepH6 has been shown to play a role in latent stage infections, as it is transcribed in latently infected lymphocytes (10).
There are two variants of HHV-6; HHV-6A and HHV-6B. HHV-6A and HHV6B share an overall nucleotide sequence identity of approximately 90%. HHV-6A has 110 ORFs, while HHV-6B has 119 ORFs. Major genomic differences between the two strains lie in the direct repeat regions near the ends of the viral genome, as well as in the proteins encoded by genes U86-U100 in the early stage of infection. The differences observed in early stage viral proteins are the result of the splicing patterns unique to each viral variant (11, 12).
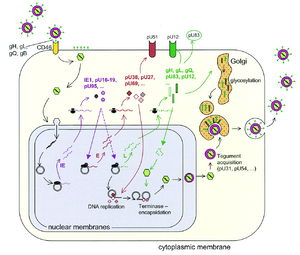
HHV-6 Replication Cycle
Epidemiology
Pathophysiology of HHV-6 Infection
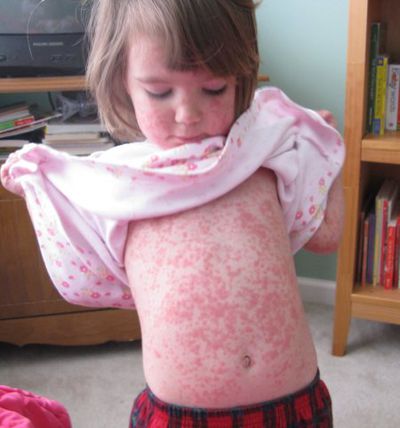
Initial Infection
Latency in Healthy Children and Adults
Reactivation in Immunosuppressed Individuals
HHV-6 and Associated Disease States
Latency through Human Chromosomal Integration
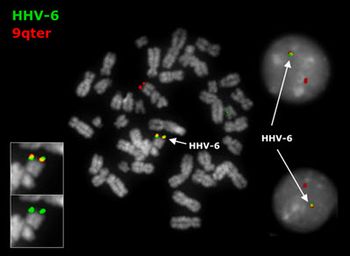
Transmission through Germ Line
Future Work
References
Edited by Kerri-Lynn Conrad, student of Joan Slonczewski for BIOL 375 Virology, 2010, Kenyon College.
