Human Metapneumovirus: Difference between revisions
No edit summary |
No edit summary |
||
| Line 12: | Line 12: | ||
<br> <br> | <br> <br> | ||
==Section 3== | ==Section 3== | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 23: | Line 21: | ||
==Vaccine Development== | ==Vaccine Development== | ||
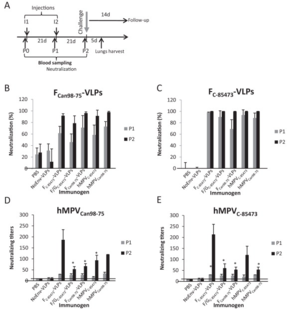
[[Image:HMPVvaccinetest.png|thumb|300px|right|Results of a VLP vaccine test performed in a mouse model. The study was performed by Lèvy <i>et al</i> in 2013, and the figure demonstrates that their vaccine tests were successful in inducing an immune response in the mice.]] | [[Image:HMPVvaccinetest.png|thumb|300px|right|Results of a VLP vaccine test performed in a mouse model. The study was performed by Lèvy <i>et al</i> in 2013, and the figure demonstrates that their vaccine tests were successful in inducing an immune response in the mice.]] | ||
<br> | <br>There has been some evidence that a vaccine for HMPV was created using virus-like particles (VLPs), rather than an attenuated form of the virus or an inactivated virus (Lèvy et al, 2013). According to this study, the VLPs are better for making a vaccine for this particular virus because, while the attenuated or inactivated vaccines can create an immune response in the test organism or patient, it does not have quite as good of a chance of lasting over a long-term time period (Lèvy et al, 2013). Because of this, the authors of this study chose to test the effects of VLPs, despite the fact that VLPs are harder to create for enveloped viruses (which all members of the paramyxovirus family are) (Lèvy et al, 2013). VLPs also have the advantage of being less dangerous to patients with suppressed or compromised immune systems because the VLP contains none of the genetic material of the virus, just a protein coat. This makes VLPs particularly applicable to the study of this virus, given that it primarily targets those who have immune systems which are not functioning properly (Schildgen et al¸2011, Lèvy et al, 2013). The purpose of the VLPs is to mimic the surface proteins found on the surface of the HMPV virion (Lèvy et al, 2013).<br> | ||
<br>HMPV has three glycoproteins which can be found on the surface of the virion: a G protein, an F protein, and an SH protein (Schildgen et al, 2011). The G protein serves the function of helping the virion attach to a host cell. The G protein is the protein which differs the most between the lineages of HMPV (Schildgen et al, 2011). The F protein facilitates the fusion of the virion membrane with the host membrane, allowing the virus to deposit its genetic material inside of the host cell (Schildgen et al, 2011). After that, the virus' genetic material is transcribed into positive-sense RNA before more proteins are made. The majority of HMPV's mechanism of reproduction has not been studied. However, it is likely that the mechanism will be similar to that of other paramyxoviruses (Schildgen et al, 2011). The third surface protein is the SH protein, the function of which has yet to be discovered (Schildgen et al, 2011). There is a significant difference in the length of the SH protein in HMPV and RSV, however, indicating that both viruses are still functional, even with their different versions of the protein. Studies were performed in which the SH protein was removed from HMPV, but it did not affect the ability of the virus to propagate or act as a pathogen (Schildgen et al, 2011).<br> | |||
<br>These three glycoproteins are mimicked on the surface of a VLP to trick the host immune system into developing a response to HMPV (Lèvy et al, 2013). The study was performed using a mouse model, where the mice were first injected with VLP for a prescribed amount of time (Lèvy et al, 2013). Blood samples were analyzed for antibody production, and at the end of the study, the mice were infected with HMPV (Lèvy et al, 2013). They were then compared to control mice (which had never been injected with VLPs) to see if there was an immune response difference (Lèvy et al, 2013). The mice which had been injected with VLPs were much more capable of resisting the HMPV infection (see figure at right). To see if the treatment would still work across different strains of HMPV, the group had two test groups, each injected with a VLP for a different strain of HMPV (Lèvy et al, 2013). These groups were then split into two again, and of each group, half was infected with the same strain as their VLP, while the other half was infected with the strain which was different from their VLP (Lèvy et al, 2013). In both cases, mice which had been exposed to VLPs fared significantly better against HMPV than mice which had not been treated with any VLPs (Lèvy et al, 2013). <br> | |||
==References== | ==References== | ||
Revision as of 01:56, 25 April 2013
Introduction
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Section 3
Symptoms
Include some current research in each topic, with at least one figure showing data.
Vaccine Development
There has been some evidence that a vaccine for HMPV was created using virus-like particles (VLPs), rather than an attenuated form of the virus or an inactivated virus (Lèvy et al, 2013). According to this study, the VLPs are better for making a vaccine for this particular virus because, while the attenuated or inactivated vaccines can create an immune response in the test organism or patient, it does not have quite as good of a chance of lasting over a long-term time period (Lèvy et al, 2013). Because of this, the authors of this study chose to test the effects of VLPs, despite the fact that VLPs are harder to create for enveloped viruses (which all members of the paramyxovirus family are) (Lèvy et al, 2013). VLPs also have the advantage of being less dangerous to patients with suppressed or compromised immune systems because the VLP contains none of the genetic material of the virus, just a protein coat. This makes VLPs particularly applicable to the study of this virus, given that it primarily targets those who have immune systems which are not functioning properly (Schildgen et al¸2011, Lèvy et al, 2013). The purpose of the VLPs is to mimic the surface proteins found on the surface of the HMPV virion (Lèvy et al, 2013).
HMPV has three glycoproteins which can be found on the surface of the virion: a G protein, an F protein, and an SH protein (Schildgen et al, 2011). The G protein serves the function of helping the virion attach to a host cell. The G protein is the protein which differs the most between the lineages of HMPV (Schildgen et al, 2011). The F protein facilitates the fusion of the virion membrane with the host membrane, allowing the virus to deposit its genetic material inside of the host cell (Schildgen et al, 2011). After that, the virus' genetic material is transcribed into positive-sense RNA before more proteins are made. The majority of HMPV's mechanism of reproduction has not been studied. However, it is likely that the mechanism will be similar to that of other paramyxoviruses (Schildgen et al, 2011). The third surface protein is the SH protein, the function of which has yet to be discovered (Schildgen et al, 2011). There is a significant difference in the length of the SH protein in HMPV and RSV, however, indicating that both viruses are still functional, even with their different versions of the protein. Studies were performed in which the SH protein was removed from HMPV, but it did not affect the ability of the virus to propagate or act as a pathogen (Schildgen et al, 2011).
These three glycoproteins are mimicked on the surface of a VLP to trick the host immune system into developing a response to HMPV (Lèvy et al, 2013). The study was performed using a mouse model, where the mice were first injected with VLP for a prescribed amount of time (Lèvy et al, 2013). Blood samples were analyzed for antibody production, and at the end of the study, the mice were infected with HMPV (Lèvy et al, 2013). They were then compared to control mice (which had never been injected with VLPs) to see if there was an immune response difference (Lèvy et al, 2013). The mice which had been injected with VLPs were much more capable of resisting the HMPV infection (see figure at right). To see if the treatment would still work across different strains of HMPV, the group had two test groups, each injected with a VLP for a different strain of HMPV (Lèvy et al, 2013). These groups were then split into two again, and of each group, half was infected with the same strain as their VLP, while the other half was infected with the strain which was different from their VLP (Lèvy et al, 2013). In both cases, mice which had been exposed to VLPs fared significantly better against HMPV than mice which had not been treated with any VLPs (Lèvy et al, 2013).
References
4. Boivin, G., Abed, Y., Pelletier, G., Ruel, L., Moisan, D., Côté, S., Peret, T. C. T., Erdman, D. D., and Anderson, L. J. (2002) Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups., The Journal of infectious diseases 186, 1330–4.
5. van den Hoogen, B. G., Bestebroer, T. M., Osterhaus, A. D. M. E., and Fouchier, R. a M. (2002) Analysis of the genomic sequence of a human metapneumovirus., Virology 295, 119–32.
6. Dokos, C., Masjosthusmann, K., Rellensmann, G., Werner, C., Schuler-Lüttmann, S., Müller, K.-M., Schiborr, M., Ehlert, K., and Groll, a H. (2013) Fatal human metapneumovirus infection following allogeneic hematopoietic stem cell transplantation., Transplant infectious disease : an official journal of the Transplantation Society 1–5.
7. Bayon-Auboyer, M.-H., Arnauld, C., Toquin, D., and Eterradossi, N. (2000) Nucleotide sequences of the F, L and G protein genes of two non-A/non-B avian pneumoviruses (APV) reveal a novel APV subgroup, J. Gen. Virol. 81, 2723–2733.
8. de Graaf, M., Herfst, S., Schrauwen, E. J. A., Choi, Y., van den Hoogen, B. G., Osterhaus, A. D. M. E., and Fouchier, R. A. M. (2008) Specificity and functional interaction of the polymerase complex proteins of human and avian metapneumoviruses., The Journal of general virology 89, 975–83.
9. Crowe, J. E. (2004) Human Metapneumovirus as a Major Cause of Human Respiratory Tract Disease, The Pediatric Infectious Disease Journal 23, S215–S221.
10. Bach, N, C., and D,; Brouard, J,; Lafay, F,; Freymuth, F,; Legrand, L,; Guillois, B,; Duhamel, J. (2004) Acute respiratory tract infections due to a human metapneumovirus in children: descriptive study and comparison with respiratory syncytial virus infections, Archives de Pediatrie : Organe Officiel de la Societe Francaise de Pediatrie 11, 212–215.
11. Peret, T. C. T., Boivin, G., Li, Y., Couillard, M., Humphrey, C., Osterhaus, A. D. M. E., Erdman, D. D., and Anderson, L. J. (2002) Characterization of human metapneumoviruses isolated from patients in North America., The Journal of infectious diseases 185, 1660–3.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2013, Kenyon College.