Influenza Vaccine
Introduction
The influenza vaccine is a highly controversial vaccine, and has several unique qualities that make it an especially interesting topic of research.
Vaccines mimic natural infections and initiate an immune response, so the body can more efficiently and affectively fight off the virus when exposed to it. The active agent of a vaccine can come in several forms, including an intact, but inactivated form of the pathogen, or a component of the pathogen that is highly immunogenic but that cannot act on its own, such as the protein coat of a virus. Exposure to one of these altered forms of the pathogen leads to the production of antibodies for the antigens of that specific virus or bacterium. The next time the body is exposed to the pathogen, it will already contain antibodies that recognize the intruder's antigens and, ideally, will be able to fight against it much more quickly.
Each year's flu vaccine is a mixture, or cocktail, of vaccines that have antigens that represent several different strains (generally 3 or 4) of the virus that the World Health Organization scientists predict will spread during that year's flu season. The predictions generally match common strains closely enough that the vaccine is fairly effective, but this is not always the case. Sometimes, a year's most common strains are unexpected ones.
The influenza virus is a rapidly evolving pathogen, which further complicates the problem. The virus evolves enough from season to season that a new dose of a modified vaccine is necessary every year, as last year's antibodies usually can not recognize this year's strain. See antigenic drift section.
Antigenic drift and new vaccine targets
Surface proteins hemagglutinin and neuraminidase are two antigens that are key players in the evolution of the influenza virus. The hemagglutinin protein helps the virus bind to and enter host cells, while the neuraminidase enzyme enables the virus to be released form the host cell as well as helps generate progeny viruses. Both proteins have specific sites that are recognized by the immune systems of the host, so these sites are under strong selection pressure. Virons with mutations in either of these proteins that allow the virus to evade attack by common antibodies are selected for by natural selection. These "fit" mutants spread to new people, and new strains evolve. This process is known as antigenic drift, as the anitigens evolve (or "drift") quickly.
Mutations occur quite often in the virus as the virus' RNA polymerase lacks a proofreading mechanism. An example of one of these mutations relates to the location of carbohydrate chains on the surface of hemagglutinin. If a mutation leads to the virus having an additional carbohydrate where the antibody binds, the antibody will no longer be effective.
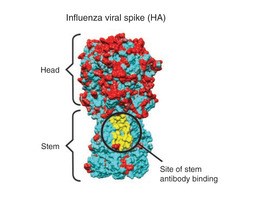
The current vaccines produce antibodies that recognize hemagglutinin protein on the outer coating of the virus, focusing on the "head" region of the molecule (see figure 2). But this "head" region is a part of the virus that is very susceptible to antigenic drift. Thus, many scientists suspect that highly conserved molecules in the influenza virus could be used as targets for a new vaccine. Several researchers are currently focusing on this possiblity. This vaccine would be universal, and would not need to be reformulated and re-administered every year. Possible targets include the hemagglutinen "stem", which is nearly identical across a wide range of strains (see figure 2), or the M2 viral membrane. A research group at NIAID's Vaccine Research Center was successful in creating a vaccine that elicits antibodies that target a specific region of the stem. They administered the experimental vaccine to animals, who were successfully protected from a vast array of strains. Whether this new target will become the key to a new, universal vaccine remains to be seen.
Risks and side affects
Include some current research, with a second image.
Conclusion
Overall text length should be at least 1,000 words (before counting references), with at least 2 images. Include at least 5 references under Reference section.
References
Goodsell, David. "Hemagglutinin." RCSB.org/pdb. RCSB Protein Data Bank, 2013. Web. 07 Nov. 2013.
Edited by [Author Name], student of Joan Slonczewski for BIOL 116 Information in Living Systems, 2013, Kenyon College.
