Megavirales Genetics from Mimivirus to Chilensis: Difference between revisions
No edit summary |
No edit summary |
||
| Line 18: | Line 18: | ||
Raoult, Didier. Reclassification of Giant Viruses Composing a Fourth Domain of Life in the New Order Megavirales ." Intervirology 2012;55:321-332. DOI: 10.1159]</ref> | Raoult, Didier. Reclassification of Giant Viruses Composing a Fourth Domain of Life in the New Order Megavirales ." Intervirology 2012;55:321-332. DOI: 10.1159]</ref> | ||
== | ==The Evolution of Nucleocytoplasmic Large DNA Viruses== | ||
<br> One of the most fascinating aspects of NCLDVs, is how they change the debate on viruses as life or not. NCLDVs share genes with all of the primary living domains; Archaea, Bacteria and Eukarya. It is especially integral that these genes are involved with the something as essential to life as DNA storage and processing(Abrahao, 2014). It begs the question how can these viruses contain so many genes required for life, and still not be considered life? In addition, it has recently been demonstrated that that Mimivirus contains its own glycosylation that leads to a 4-amino-4 6-dideoxy-D-glucose, called viosamine. It is also possible that this pathway could have evolved and changed to become similar eukaryotic glycosylation pathways. The presence of this sugar is observed even without the presence of regular amoeba host, suggesting the potential for autonomous, or semi-autonomous growth of Mimivirus(Piacente,2011). Environmentally the role of aquatic NCLDVs has also been imagined. With the suggestion being that the role of Mimiviruses perhaps was to lyse prokaryotes or eukaryotes and release carbon containing compounds, or other nutrients to increase the presence of microbes or any other systems or animals that might depend on this cell lysis. When it comes to considering virus-early or virus-late hypotheses, unfortunately there is not a clear cut answer in differing phylogenetic trees. Analysis of highly conserved genes suggest that both hypotheses are possible, Eukaryotes may share an ancestor for all NCLDVs, until this ancestor eventually diverged from eukaryotic life. It is also possible that Eukaryotes were already a distinguished group and played host to the ancestor of modern day NCLDVs, until their differentiation was mediated by horizontal virus transfer(Yutin and Koonin, 2010). | |||
Include some current research, with at least one figure showing data.<br> | Include some current research, with at least one figure showing data.<br> | ||
<br> | <br> | ||
Every point of information REQUIRES CITATION using the citation tool shown above. | Every point of information REQUIRES CITATION using the citation tool shown above. | ||
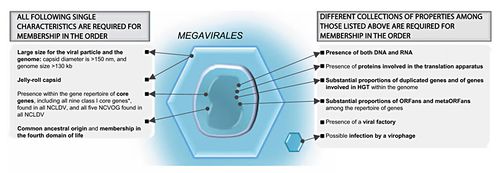
==Support For Megavirales as a Fourth Domain== | |||
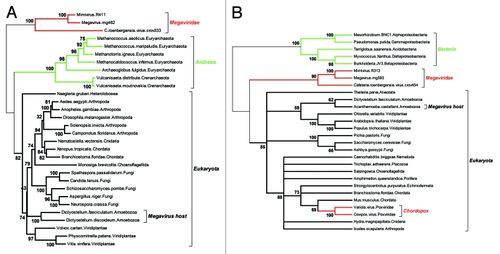
<br>The classification of NCLDVs as a defining phylogenetic group, or the establishment of Megavirales as a proposed new order is one that does not go unchallenged in the face of certain objections to phylogenies created from viral DNA. This is for two main reasons: Viruses evolve at an extremely rapid rate meaning that consistency will be difficult to maintain from one genetic analysis to the next, within a small set of constantly changing genes. The second reason is that viruses are liable to engage regularly in horizontal gene transfer with the host muddying up clear relationships between species or patterns of descent. However, in the case of NCLDVs these objections are less applicable. These double-stranded DNA viruses do not evolve at a rate comparable to smaller viruses with homologs that are significantly conserved, therefore we might expect the significant difference between NCLDVs to be a result of divergence that has occurred over a significant amount of time, suggesting a common ancestor for NCLDVs. The idea that horizontal gene transfer occurs regularly is also challenged in Megaviruses, due to their ability to rely completely on the cellular processes of the host. Therefore, there is not much evolutionary incentive for them to retain host genomes in NCLDV populations, in fact the pressure would suggest more static gene populations, or further reductive evolution, barring some unforeseen environmental shift. There is also not genetic evidence for clear-cut examples of gene transfer that has occurred between Eukaryotic hosts and their associated NCLDVs. For example, the open reading frames for non-homologous genes between Megavirus and MImivirus are 80% different, much higher than would be expected in the case of horizontal gene transfer. However, certain sets of genes (described further in gene product section) are sufficiently conserved in order to facilitate and form an adequate phylogeny (Legendre, 2012). To build a phylogeny, the Legendre paper used two specific genes that had excellent conservation in NCLDV’s and that were necessary for DNA replication. The first being a ribonucleoside diphosphate reductase sub-unit and a DNA replication clamp loader called factor C. This analysis illustrates that these genes show clear distinction from Archaea and Eukaryotes, showing little, or no overlapping affinities between these orders. Said another way, Megavirales as a proposed order, appears just as separate from Eukaryotes as Archaea are. Also up for speculation is whether or not Megavirales are at the root of the Eukaryotic domain as in Figure 2A, or are not evolutionary pre-cursors to Eukaryotes as in Fig. 2B. | |||
[[Image:Megavirales Domain.jpg|thumb|500px|right|Figure 3. Classification and Grouping of Megavirales organisms(http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3291303/bin/cib-5-102-g2.jpg]] | |||
Include some current research, with at least one figure showing data.<br> | |||
However, phylogeny based on these two gene regions as in figure 3. are not necessarily the only kind a researcher could choose to build. In fact other work using DNA ligase as the primary gene to build phylogeny, has not seen such a clear cut relationship(Yutin and Koonin, 2012). When using DNA ligase Yutin and Koonin found that a presence-absence analysis of a particular kind of DNA ligase showed clear progression and separtion of an ancestral and current form. However, when supplementing the very same analysis with other DNA ligase genes, the likely ancestor became the more recent ligase and the other ligase was projected as the ancestor. This shows that although there is obvious support for a common ancestor for NCLDVs, not all phylogenies will place genes chronologically in the same order. Some phylogenies are not even able to statistically separate or group the proposed NCLDVs with significance. It all depends on the genes you choose to analyze them and weighting certain genes as more or less important is extremely complicated and difficult(Yutin and Koonin, 2012). Ultimately, this work suggests the opposite conclusions of other papers, and hints at the possibility of independent acquisition of genes by NCLDVs from cellular hosts, as a way of explaining difficulties in consistently classifying these different large viruses. Support for Megavirales as a new life classification is also slightly contested by the fact that Poxviridae and Ascoviridae, are both NCLDVs that don’t feature icosahedral capsids, instead they have brick-shaped allantoid capsids. However, in Poxviridae organisms it is thought that this feature is a derived one, due to the fact that during morphogenesis icosahedral form is expressed as intermediate form, and the D13 protein is still transcribed and translated during this developmental stage(Colson, 2013). | |||
==Section 3== | ==Section 3== | ||
| Line 46: | Line 36: | ||
<br> | <br> | ||
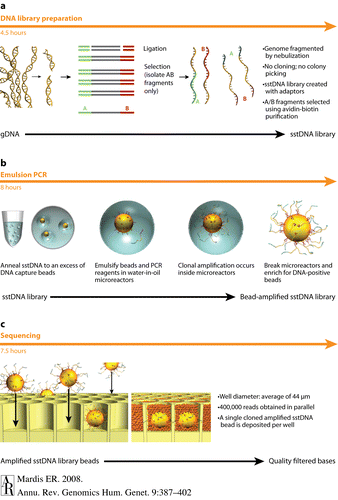
[[Image:Roche sequencing.gif|thumb|500px|right|A summary and visualization of Roche-454 Pyrosequencing methods(http://www.annualreviews.org/na101/home/literatum/publisher/ar/journals/content/genom/2008/genom.2008.9.issue-1/annurev.genom.9.081307.164359/production/images/medium/gg090387.f1.gif]] | [[Image:Roche sequencing.gif|thumb|500px|right|Figure 4: A summary and visualization of Roche-454 Pyrosequencing methods(http://www.annualreviews.org/na101/home/literatum/publisher/ar/journals/content/genom/2008/genom.2008.9.issue-1/annurev.genom.9.081307.164359/production/images/medium/gg090387.f1.gif]] | ||
==Section 4== | ==Section 4== | ||
[[Image:Illumina sequencing.gif|thumb|500px|right|The Mimivirus replication cyclee visualized with Transmission Electron Microscopy(http://www.annualreviews.org/na101/home/literatum/publisher/ar/journals/content/genom/2008/genom.2008.9.issue-1/annurev.genom.9.081307.164359/production/images/medium/gg090387.f2.gif]] | [[Image:Illumina sequencing.gif|thumb|500px|right|Figure 5: The Mimivirus replication cyclee visualized with Transmission Electron Microscopy(http://www.annualreviews.org/na101/home/literatum/publisher/ar/journals/content/genom/2008/genom.2008.9.issue-1/annurev.genom.9.081307.164359/production/images/medium/gg090387.f2.gif]] | ||
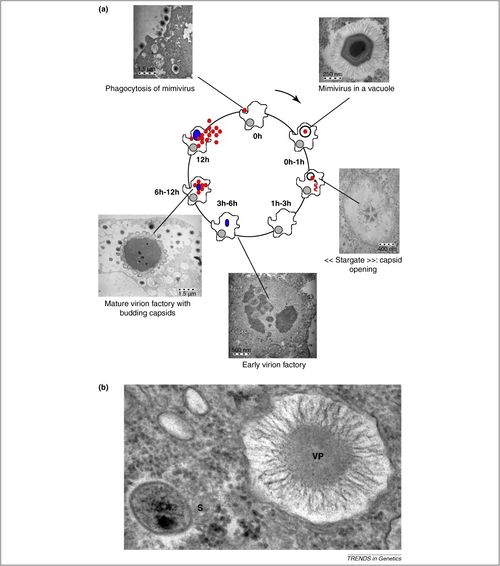
[[Image:Mimivirus replication.jpg|thumb|500px|right|The Mimivirus replication cycle visualized with Transmission Electron Microscopy(http://ars.els-cdn.com/content/image/1-s2.0-S0168952510001460-gr1.jpg]] | [[Image:Mimivirus replication.jpg|thumb|500px|right|Figure 6: The Mimivirus replication cycle visualized with Transmission Electron Microscopy(http://ars.els-cdn.com/content/image/1-s2.0-S0168952510001460-gr1.jpg]] | ||
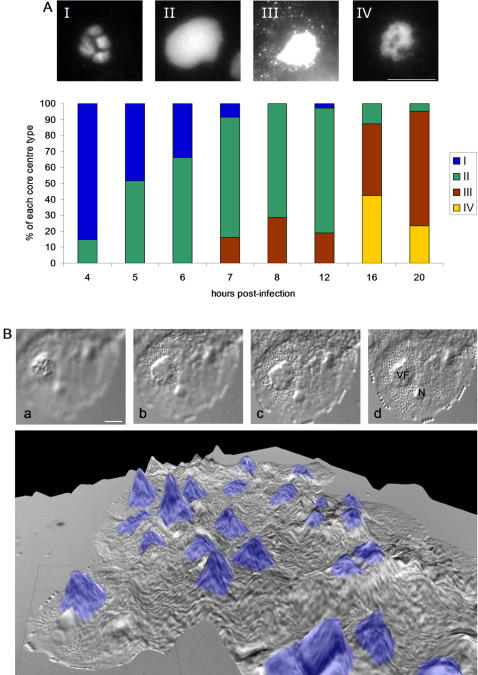
[[Image:Viral Factory.jpg|thumb|500px|right|Visualization of Volcanic-Like Viral Factories with DIC and DAPI staining(http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1828621/bin/pone.0000328.g005.jpg]] | [[Image:Viral Factory.jpg|thumb|500px|right|Figure 7: Visualization of Volcanic-Like Viral Factories with DIC and DAPI staining(http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1828621/bin/pone.0000328.g005.jpg]] | ||
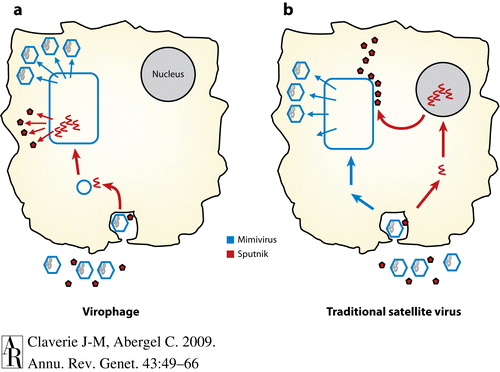
[[Image:Virophage Replication.gif|thumb|500px|left Virophage replication using NCLDV viral factories(http://www.annualreviews.org/na101/home/literatum/publisher/ar/journals/content/genet/2009/genet.2009.43.issue-1/annurev-genet-102108-134255/production/images/medium/ge430049.f6.gif]] | [[Image:Virophage Replication.gif|thumb|500px|Figure 8: left Virophage replication using NCLDV viral factories(http://www.annualreviews.org/na101/home/literatum/publisher/ar/journals/content/genet/2009/genet.2009.43.issue-1/annurev-genet-102108-134255/production/images/medium/ge430049.f6.gif]] | ||
==Conclusion== | ==Conclusion== | ||
Revision as of 19:38, 27 April 2016
By Adama Berndt
Introduction
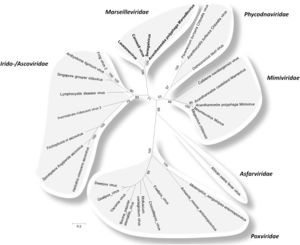
The evolutionary history of viruses is an unclear and frequent topic of speculation. Megaviruses are especially unique, as their size and structure are reminiscent of bacterial cells, but their function is similar to that of a classical virus(Abrahao, 2014). Although knowledge of large viruses dates back to the 1970’s, interest in megaviruses expanded during 2003 with the sequencing of Mimivirus; now one of the most well characterized Nucleocytoplasmic Large DNA Viruses(NCLDVs)(Colson, 2012). There are 6 official recognized classes of NCLDVs: Poxviridae, Asfarviridae, Iridoviridae, Ascoviridae, Phycoviridae, Mimiviridae(Colson, 2013; Fig 2.). In addition to these classes Megaviridae is proposed as a seventh class(Yutin and Koonin, 2012). One of the most remarkable recurring traits of these microbes is their large genome size, which at approximately one million base pairs is bigger than some bacterial genomes. These base pairs code for over a thousand genes and this genetic material is surrounded by pseudo-icosahedral protein capsids, similar to a Herpes virus(Legendre, 2012)(Fig. 1.). The taxonomy of viruses as DNA containing microbes has long been considered from many angles. Of special consideration is the status of NCLDVs as living or non-living organisms. From an evolutionary perspective, there are two major sides to this debate, the “virus-early” hypothesis the “virus-late’ hypothesis. The virus-early stance argues that viruses originated before modern cells and the virus-late stance speculates that viruses are the result of a reductive evolution, characterized by an increased benefit from reliance on host metabolism, which propelled the loss of primary cellular machinery. The other potential mechanism of the virus-late camp imagines the escape of proteins and genetic information from cells, that coupled to form viruses(Moreira, 2008). If viruses were formed spontaneously from released cell genetic materials and proteins, me might expect to see some formation of new rudimentary viruses today. The genetic information obtained from Mimivirus has provided extra fuel for speculation in this debate, specifically that Mimivirus contains the first evidence of a virus containing genes for transcription and translation(Raoult, 2007). The full phylogenetic analysis conducted by Raoult and colleagues found several Eukaryotic protein homologues in Mimivirus, and support for Mimivirus as a sister group to Eukaryotes and therefore a new branch of life that would include, Eukaryotes, Prokaryotes, Archaea and Megavirales.
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
Introduce the topic of your paper. What is your research question? What experiments have addressed your question? Applications for medicine and/or environment?
A citation code consists of a hyperlinked reference within "ref" begin and end codes.
[1]
The Evolution of Nucleocytoplasmic Large DNA Viruses
One of the most fascinating aspects of NCLDVs, is how they change the debate on viruses as life or not. NCLDVs share genes with all of the primary living domains; Archaea, Bacteria and Eukarya. It is especially integral that these genes are involved with the something as essential to life as DNA storage and processing(Abrahao, 2014). It begs the question how can these viruses contain so many genes required for life, and still not be considered life? In addition, it has recently been demonstrated that that Mimivirus contains its own glycosylation that leads to a 4-amino-4 6-dideoxy-D-glucose, called viosamine. It is also possible that this pathway could have evolved and changed to become similar eukaryotic glycosylation pathways. The presence of this sugar is observed even without the presence of regular amoeba host, suggesting the potential for autonomous, or semi-autonomous growth of Mimivirus(Piacente,2011). Environmentally the role of aquatic NCLDVs has also been imagined. With the suggestion being that the role of Mimiviruses perhaps was to lyse prokaryotes or eukaryotes and release carbon containing compounds, or other nutrients to increase the presence of microbes or any other systems or animals that might depend on this cell lysis. When it comes to considering virus-early or virus-late hypotheses, unfortunately there is not a clear cut answer in differing phylogenetic trees. Analysis of highly conserved genes suggest that both hypotheses are possible, Eukaryotes may share an ancestor for all NCLDVs, until this ancestor eventually diverged from eukaryotic life. It is also possible that Eukaryotes were already a distinguished group and played host to the ancestor of modern day NCLDVs, until their differentiation was mediated by horizontal virus transfer(Yutin and Koonin, 2010).
Include some current research, with at least one figure showing data.
Every point of information REQUIRES CITATION using the citation tool shown above.
Support For Megavirales as a Fourth Domain
The classification of NCLDVs as a defining phylogenetic group, or the establishment of Megavirales as a proposed new order is one that does not go unchallenged in the face of certain objections to phylogenies created from viral DNA. This is for two main reasons: Viruses evolve at an extremely rapid rate meaning that consistency will be difficult to maintain from one genetic analysis to the next, within a small set of constantly changing genes. The second reason is that viruses are liable to engage regularly in horizontal gene transfer with the host muddying up clear relationships between species or patterns of descent. However, in the case of NCLDVs these objections are less applicable. These double-stranded DNA viruses do not evolve at a rate comparable to smaller viruses with homologs that are significantly conserved, therefore we might expect the significant difference between NCLDVs to be a result of divergence that has occurred over a significant amount of time, suggesting a common ancestor for NCLDVs. The idea that horizontal gene transfer occurs regularly is also challenged in Megaviruses, due to their ability to rely completely on the cellular processes of the host. Therefore, there is not much evolutionary incentive for them to retain host genomes in NCLDV populations, in fact the pressure would suggest more static gene populations, or further reductive evolution, barring some unforeseen environmental shift. There is also not genetic evidence for clear-cut examples of gene transfer that has occurred between Eukaryotic hosts and their associated NCLDVs. For example, the open reading frames for non-homologous genes between Megavirus and MImivirus are 80% different, much higher than would be expected in the case of horizontal gene transfer. However, certain sets of genes (described further in gene product section) are sufficiently conserved in order to facilitate and form an adequate phylogeny (Legendre, 2012). To build a phylogeny, the Legendre paper used two specific genes that had excellent conservation in NCLDV’s and that were necessary for DNA replication. The first being a ribonucleoside diphosphate reductase sub-unit and a DNA replication clamp loader called factor C. This analysis illustrates that these genes show clear distinction from Archaea and Eukaryotes, showing little, or no overlapping affinities between these orders. Said another way, Megavirales as a proposed order, appears just as separate from Eukaryotes as Archaea are. Also up for speculation is whether or not Megavirales are at the root of the Eukaryotic domain as in Figure 2A, or are not evolutionary pre-cursors to Eukaryotes as in Fig. 2B.
Include some current research, with at least one figure showing data.
However, phylogeny based on these two gene regions as in figure 3. are not necessarily the only kind a researcher could choose to build. In fact other work using DNA ligase as the primary gene to build phylogeny, has not seen such a clear cut relationship(Yutin and Koonin, 2012). When using DNA ligase Yutin and Koonin found that a presence-absence analysis of a particular kind of DNA ligase showed clear progression and separtion of an ancestral and current form. However, when supplementing the very same analysis with other DNA ligase genes, the likely ancestor became the more recent ligase and the other ligase was projected as the ancestor. This shows that although there is obvious support for a common ancestor for NCLDVs, not all phylogenies will place genes chronologically in the same order. Some phylogenies are not even able to statistically separate or group the proposed NCLDVs with significance. It all depends on the genes you choose to analyze them and weighting certain genes as more or less important is extremely complicated and difficult(Yutin and Koonin, 2012). Ultimately, this work suggests the opposite conclusions of other papers, and hints at the possibility of independent acquisition of genes by NCLDVs from cellular hosts, as a way of explaining difficulties in consistently classifying these different large viruses. Support for Megavirales as a new life classification is also slightly contested by the fact that Poxviridae and Ascoviridae, are both NCLDVs that don’t feature icosahedral capsids, instead they have brick-shaped allantoid capsids. However, in Poxviridae organisms it is thought that this feature is a derived one, due to the fact that during morphogenesis icosahedral form is expressed as intermediate form, and the D13 protein is still transcribed and translated during this developmental stage(Colson, 2013).
Section 3
Include some current research, with at least one figure showing data.
Section 4

Conclusion
References
- ↑ [http://www.karger.com/Article/Fulltext/336562 Colson, Phillipe. Lamballerie, Xavier de. Fournous, Ghislain. Raoult, Didier. Reclassification of Giant Viruses Composing a Fourth Domain of Life in the New Order Megavirales ." Intervirology 2012;55:321-332. DOI: 10.1159]
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2016, Kenyon College.
