Metabolic Processes in Rhodopseudomonas palustris
Introduction
By A. Pearlman
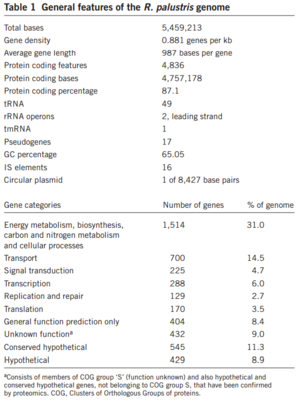
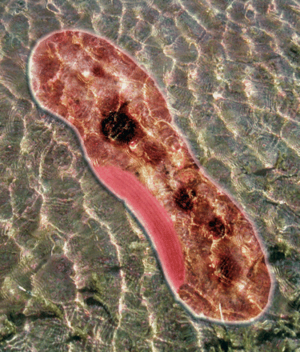
Rhodopseudomonas palustris, recognizable by its purple color, is a gram-negative alpha proteobacterium commonly found in pond and river water as well as the surface of soil and rock. They do not typically form any sort of biofilm. The species is capable of a remarkably wide host of metabolic processes, including aerobic respiration, anxoygenic photosynthesis (Harwood Lab), carbon fixation, nitrogen fixation, and extracellular electron transfer. These modes of metabolism are not all active at once, and are triggered in response to certain environmental cues. In order to support all these processes efficiently, much of its 5.5 million bp genome is devoted to regulation, signaling, and transport (Figure 1). Furthermore, its genome has an unusually smaller number of redundancies (theoretically to maximize the amount of varying information that can be stored) and does not appear to bear the products of many phage genome integrations or horizontal gene transfers, further begging the question of how it was able to evolve so many different metabolisms (Larimer et. al 2004).
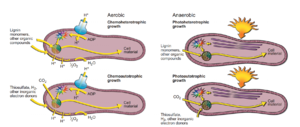
R. palustris has two potential reproduction cycles that it may follow, depending on the environmental conditions. Much of the energy used in the reproduction process is derived from light-based metabolism, and is possible in anaerobic and aerobic conditions. In the presence of oxygen, R. palustris will generally undergo budding, resulting in two daughter cells (Figure). One of these daughter cells will not be motile and will possess a stalk (derived from the original parent cell), while the other will be motile and is classified as a "swarmer." Under hypoxic conditions, R. palustris cells will develop "intracytoplasmic membranes vesicles" that eventually differentiate into the daughter cells. It should be noted that some degree of light is necessary for this process. (Achanta)
The aerobic reproduction proceeds via 4 primary steps:
- First, a bud begins to form opposite of the parent cell's flagellum. This bud is tubular in shape, and extends to up to two times the size of the parent body. The tube was shown to be relatively elastic, and was able to bend around nearby detritus if necessary.
- In the second stage, the far end of the tube-like bud grows to a more spherical shape as its organelles develop more.
- After sufficient growth of the bud, the parent and daughter cells divide in an asymmetric fashion, with the parent cell retaining most of the tube-like appendage. As the division happens, the daughter cell is able to move/swim away from the parent. The parent cell, however, is no longer motile at all.
- In the fourth stage, the daughter cell matures by growing in length to the shape typical of R. palustris and eventually appears identical to the parent cell after enough development, save that it possesses motile ability.
The parent cell still possesses the ability to reproduce normally, and has actually been shown to reproduce faster than before, as it has already developed the tube-like structure where the bud forms (its reproduction time has been shown to be 210 minutes on average). The daughter cells can reproduce as well, but will have to completely develop the budding structure before that is possible (resulting in a reproduction time of about 221 minutes total).
(Whittenbury & McLee 1967)
Genomics
Include some current research, with at least one figure showing data.
Since R. palustris supports such a variety of metabolic systems, much of its genome is accordingly devoted to metabolism. So far, some of the metabolism-related gene regions that allow for oxidation of thiosulfate ions, gaseous elemental hydrogen, carbon monoxide, and carbon dioxide have been found, sequenced, and analyzed. Furthermore, certain regions have been identified that allow for R. palustris to break down a host of organic molecules, such as lignin and a variety of fatty acids. Interestingly, there is code for two separate enzyme systems, each of which appears to confer the same catabolic abilities. As of yet, it is not known why this apparent redundancy exists in a species of microbe that is quite constrained for space in its genome, nor has it been determined if different conditions will trigger one of these systems of the other. Other key features of the R. palustris genome include code for 4 distinct oxidase genes, which are stimulated into expression in aerobic conditions. (Larimer 2004)
Species such as R. palustris are quite common in water sanitization facilities, especially in the presence of sunlight, because its metabolism allows it to degrade and utilize much of the waste present in the water. As such, it was hypothesized that a significant portion of the genome would be dedicated to these metabolic processes. The prediction has proved accurate, and the species has been shown to possess the ability to utilize many more classes of organic compounds than were previously imagined. (Larimer 2004)
As R. palustris is capable of the Calvin Cycle, enzymes related to this process were, of course, predicted to be coded for in the genome. It turns out that R. palustris is actually the only living organism with two ruBisCo-like proteins, or RLPs, coded for in its genes. Not much has been discovered concerning the purpose of this arrangement, but it is theorized that they are important for metabolism of sulfur compounds. (Larimer 2004)