Mouth
Introduction
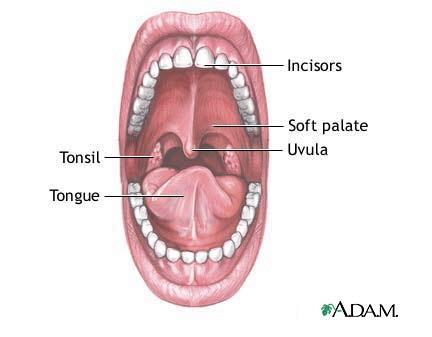
The dark, wet, and warm environment of the mouth, with the occasional meal running through it, makes it an excellent niche for microbes to live. Over the past 40 years, scientists have been arduously working to discover the over 500 different species of bacteria in and around the mouth known today. The mouth is comprised of an oral cavity, which includes the teeth and gums, surrounded by the lips, cheeks, tongue, palate, and throat (Figure 1). Each of these habitats offers differing environmental conditions (Figure 2), and as such, is colonized by a different microbial flora. The oral environment is constantly in flux. From birth to around age 12, when the permanent dentition is complete, the local oral conditions are continuously changing as teeth are shed and new ones erupt (The Normal Microbial Flora of Man). In addition, environmental factors such as, nutrition, diet, hygiene, smoking, dehydration, and even stress, alter the ecological conditions of mouth. Saliva covers all surfaces and serves various important functions, mechanical and nutritional, (digestive, swallowing, cleansing, lubricative, bactericidal, and excretory) in the oral cavity. It is comprised of various proteins and glycoprotein, of which the main constituents include salivary mucins (approximately 25% of saliva), amylase, IgA, and lysozyme. The typical resting pH (6.5-6.9) is slightly more acidic than stimulating pH (7.0-7.5), but often varies depending on the secretion rate. (Indigenous Microbiota of the oral cavity). While salivary flora does not necessarily represent the microbial composition of the different components of the mouth, it does impact which microbes can live within the oral cavity, and has recently been the target of research in early disease detection (link to current research).
Teeth
Description
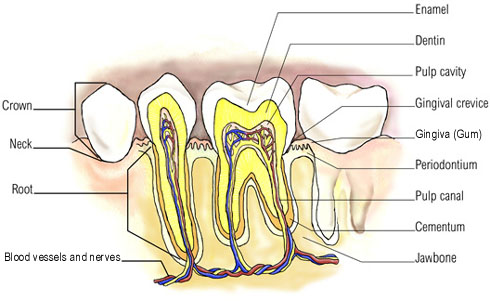
The second largest biomass of bacteria within the mouth is found on the surface of the tooth enamel, with an estimation of 10^11 organisms per gram wet weight (B1). This large and diverse population of bacteria is able to reside on the enamel because they form well organized communities known as a biofilm or more commonly supragingival plaque. “Biofilms are analogous to the planet Earth, where the properties of the latter as a whole are determined by the interactions of all of the residents as well as interactions of the populations with inanimate supporting structures” (B2). The bacteria within the mouth not only have to communicate with all the different species living within the biofilm, but they must obtain a strong adherence to a surface in order to not be washed away by saliva.
The teeth are one of the favored surfaces by bacteria not only because it is a hard surface to adhere to, but the saliva and surface location keeps the oral cavity at warm conditions of 35-36°C, moist, at a neutral to slightly acidic pH due to a bicarbonate ion buffer and bacterial metabolites, and provides nutrients for the bacteria (B3,B4). The bacteria can also obtain most of its nutritional needs from exogenous nutrients introduced by the host’s diet. These nutrients provide carbohydrates, proteins, and glycoproteins needed for the growth (B3). The two most common sites where supragingival plaque is found are the approximal and fissure surfaces of the enamel. The approximal surface, between the teeth, is protected from abrasion from salivary flow and oral hygiene, has a rich supply of nutrients from saliva, the host’s diet, microbial products, and gingival crevicular fluid, has low oxygen levels, and has a low redox potential; which allows for predominantly anaerobic bacteria (B3,B4). The fissures, located on the crowns of the teeth, are more exposed to abrasion, often get food particles packed into crevices, has a high oxygen level, and has a high redox potential, so more diverse range of bacteria reside here (B4).
Who lives there?
Which microbes are present?
The mouth conditions change throughout the day depending on saliva flow, food, and oxygen levels. This fluctuation of oral conditions causes the population of the biofilm within the mouth to also change because it will affect the bacteria-bacteria interactions. Although studies show that the common microflora on the enamel are constantly changing due to environmental conditions and location, the most common bacteria that are found in the supragingival plaque are: gram positive cocci (Streptococcus mitis, S. oralis, S. sanguis, S. mutans, S. gordonii, and staphylococcus aureus, staphylococcus epidermidis); gram negative cocci and rods (Veillonella spp., Neisseria sicca, and Fusobacteria); and gram positive rods and filaments (Actinomyces spp., Corynebacterium spp., lactobacilli, and Prevotella spp.) (B5, B6, B2). These prominent bacteria are responsible for the plaque formation due to their interactions with each other and the tooth surface.
Do the microbes that are present interact with each other?
Pellicle formation
In order for the bacteria to form a biofilm first adhere to the tooth’s surface. In the mouth, the tooth forms its own layer of protection known as the acquired pellicle. While protection is its main purpose, it also serves as lubrication for the enamel surface and acts as a semi-permeable membrane (B7). The pellicle is composed mostly from components of saliva such as proteins, lipids, and carbohydrates, and is never larger than one micrometer thick (B1). Like supragingival plaque, the acquired pellicle’s composition is believed to differ depending on the region of the mouth because the flow of saliva and the biopolymers that makes up the saliva.
The dental enamel is made of inorganic minerals which mostly consist of calcium and phosphate. The proteins of the saliva bind to the most common calcium phosphates, hydroxyapatite, through an electrostatic interaction. Some of these proteins are the phosphoproteins: statherin, histatin, and proline-rich proteins (PRPs). Although these have been commonly found to form the initial layer of the acquire pellicle, studies have shown that there may be other interactions, such as Van der Waal forces and hydrophobic interactions, that also play an important role in protein-enamel interactions. The second stage of pellicle formation involves protein-protein interactions to build up the layers. Like the biofilm, the pellicle is constantly undergoing modifications depending on the host enzymes and microbial metabolites within the mouth.
The pellicle is able to form a protective layer around the enamel surface because of its many antimicrobial components. Some of these components are: lysozyme, Cystatin, sIgA, and Histatin (B7). Although the pellicle is initially made to protect the enamel surface, its components also makes a ‘good’ surface for adherence. The early colonizers of the biofilm have receptors that have a high affinity for the PRPs, statherin proteins, α-amylase, and glycoproteins on the pellicle (B8, B9).
Biofilm formation
The bacteria that live on the tooth surface form multi-specie communities known as a biofilm. A biofilm can be called a community because the bacteria work together to organize themselves in such a way to maintain homeostasis by resisting changes in their environment. Biofilms can resist antimicrobial agents, bacterial enzymes, pH changes to some degree due to the different components of the overall structure of the biofilm and individual contributions each species of bacteria contributes (B6, B3). Formation of the pellicle on the tooth’s surface gives the other bacteria a face to bind to and eventually leads to the formation of a biofilm. The interactions that allow the bacteria to bind to the pellicle are split up into two stages. When the microbe is still some distance to the pellicle-coated enamel, long-rang physicochemical forces allow for weak, reversible attachment. As the microbe comes closer, molecules called adhesion, on the microbe’s surface, form strong, irreversible interactions with the acquired pellicle. Adhesins are molecular components of the binding structures: pili, fibrils and fimbriae. This process happens through strong-range sterochemical forces. Most oral bacteria have more than one type of adhesin on its surface to allow it to be able to bind to different pellicle sites (B12, B7).
The first bacteria that come in contact with the pellicle surface are known as primary colonizers. These bacteria must be able to withstand high oxygen concentrations and abrasion (B6). Streptococci make up 80% of the primary colonizers, more specifically S. oralis, S. sanguis, and S. mitis (B7). As these attach themselves, they in turn provide new surface receptors that adhesin proteins of later colonizers can bind. Some of the most common secondary colonizers are Veilonella spp., Actinomyces naselundii, Prevotella spp., and Neisseria. These earlier colonizers deplete the oxygen within the local environment, and allow for anaerobes to bind to the biofilm (B3, B10). A notable secondary colonizer is Fusobacterium nucleatum which is important base for tertiary and further layers because it forms a bridge between the early and late colonizers. It has receptors that almost all oral bacteria can bind to. As more bacteria attach themselves and multiply they tend to grow upwards in a columnar fashion, perpendicular to the tooth’s surface (B1, B9). This dictates the overall population and spatiotemporal formation of the biofilm.
The biofilm is able to withstand environmental pressures because they form a protective outer layer. As the Streptococci and other primary colonizers begin their attachment they utilize sugars mainly sucrose to start polysaccharide production. The polysaccharides are synthesized using enzymes called glycosyltransferases which are located on the cell’s surface. When sucrose is broken down it has two components fructose and glucose, further enzymes and bacterial reactions creates bonds between multiple fructose molecules forming fructan. The same is done with glucose which is called glucan. Glucan and fructan are than brought into the cell and can later be broken down and used as an energy source (B13). The microbes also use these polysaccharides as a glue-like substance to further secure their attachment to the pellicle-coated enamel and also to form one of the staple components in any mature biofilm, the extracellular matrix (B1, B12). The matrix provides structural support that allows the microbes to attach in a columnar fashion and also gives the biofilm a strong attachment to the pellicle to prevent it from washing away. One would tend to believe that the extracellular matrix is a solid structure surrounding the cell, but on the contrary it is very open with channels and voids running along it. This is to help the biofilm retain nutrients and water; it also allows them to pass these nutrients in between the channels to other microbes in the community.
Cell-to-Cell Communication
However, the biofilm is not just a simple community where bacteria can easily bind to nascent surfaces to build upon. The formation of the biofilm is more complex, and involves many interactions between genetically similar and distant species. The biofilm must maintain homeostasis, constant environmental conditions, in order to maintain a healthy mouth. The commensal bacteria compete with their neighbors in order to dictate the conditions of the biofilm (B3). Through antagonistic and synergistic interactions, the bacteria exhibit the following communication possibilities: coaggregation and coadhesion, metabolic products, gene expression and transformation, quorum sensing, as well as cell-cell signaling are the many ways the bacteria communicate (B6, B9). Studies are still ongoing to fully understand how the oral microflora is able to from such diverse and complex communities, but it is agreed that communication is essential.
For the early colonizers, who withstand many factors from both the environment and their neighbors, they must have a variety of different receptors and adhesins to perform all their functions. The Streptococcus genus is always one the first and most popular bacteria found at the foundation of the biofilm because they are one of the few bacteria that exhibit intergeneric coaggregation, the ability to bind to both the host and other bacteria. For example, S. gordonii binds to multiple receptors on the acquired pellicle as well as with its neighbors. It binds to salivary agglutinin glycoproteins by the receptor SspA/B, to α-amaylase by AbpA, α-2-3-linked sialic acid termini of O-linked oligosaccharides of glycoconjugates by the protein Has, and also recognizes the Pro-Gln dipeptide of proline-rich proteins (B11). Other early colonizers like actinomyces, capnoyctophagae, haemophili, prevotellae, propionibacteria, and veillonellae also recognize the components in the acquired pellicle through specific cell surface adherence proteins (B11). However, studies have shown that individual attachment and growth of monospecies can survive within the mouth.
Bacteria-bacteria interactions increase their affinity for a communal lifestyle. Without coaggregation and coadhesion, the multi-specie biofilms would not be able to form. Coaggregation is the recognition and communication between bacteria in suspension that will clump together to form an aggregate that can bind to the biofilm; while coadhesion is the adhesion of individual bacteria cell in suspension with a cell that is already a member of the biofilm. These interactions are mediated by complementary protein-adhesin and saccharide-receptors (B10). Coaggregation between streptococci and actinomcyes, initial colonizers, help them bind to the acquired pellicle as well as manipulate spatiotemperoral development of plaque. Cell-cell interactions are formed through lectin-like receptors; which involves a protein adhesion recognizing the streptococcal receptor polyscahharide (RPS). Streptococci can interact between each other; seen when Gn RPS on S. oralis binds with the protein adhesion DL1 on S. gordonii. Coaggregations also occur between different genuses; for example, Gn and G RPS on streptococci is recognized by the protein adhsin type 2 fimbriae on actinomyces (B15). Without coaggregation, both S. oralis and A. naelundii would not have a high enough affinity to form even a monoculture on the acquired pellicle, but by their combined metabolic products are able to use the salivary components (B2)
Coaggregation and further addition of later colonizers to the biofilm are dictated by the bacteria’s nutritional and environmental needs (B6). The earlier colonizers are essential to the formation of the biofilm because they change the environmental conditions for the next layer of bacteria to adhere. After S. gordonii has adhered to the acquired pellicle, its interaction with the saliva results in the gene expression of DL1 which encodes for the antigen I/II adhesions, specifically SspA/B. The presence of these proteins promotes its ability to bind to salivary agglutinin as well as the coaggregation of S. gordonii with the other colonizers Actinomyces and P. gingivalis (B8, B9).
Throughout the biofilm, the overall structure is dictated by antagonistic and synergistic interactions between the bacteria. The composition of the biofilm has a basic organizational method, but the actual percentage of each bacterium present differs between individuals most likely because of interactions that occur. A. naeslundii coadheres to S. oralis in an interaction that promotes the growth for both partners. However, since S. gordonii competes for the same nutrients, if A. naeslundii-S. oralis are present, there is a decrease in the S. gordonii population (B10). The initial colonizers will determine which colonizers will be able to adhere later. For example, if S. mutans is present on the biofilm first, its production of hydrogen peroxide will inhibit the binding and growth of S. mutans since the anaerobe cannot destroy this reactive oxygen intermediate. If S. mutans binds first, its produces bacteriocins, Mutacin I and IV, which will prevent S. sanguinis from binding since it cannot tolerate this toxin (B2, B14). Some interactions promote synergistic behavior. Some bacteria act as a bridge between bacteria that would not normally coadhere or coaggregate with each other (B9, B2)
One of the most important advantages that bacteria have by living in a biofilm, is that the bacteria are able to withstand more environmental pressures because if the biofilm structure and communication. The bacteria send diffusible signals to their neighbors in order to communicate. One method, quorum sensing, involves the signaling molecule, autoinducer-2 (AI-2) produced from the gene LuxS. This is frequently found in both gram-positive and gram-negative bacteria and is suggested to have a wide range of functions for the community. In biofilms that do not have properly working AI-2 molcules, or those similar, are not able to maintain homeostasis. S. mutans specifically use a competence stimulating peptide (CPS), and the production of this molecule increases its ability to perform transformation by 10-600 times more frequently (B16,B8). Gene exchange allows the inhabitants the ability share. If a cell lyses, the free DNA can be up taken into a competent cell and incorporated into its own genome, can be used as a nutrient source, or just as a means of communication (B9). The transfer of conjugative transposons, jumping genes, can allow for horizontal gene transfer, between different geniuses. The bacteria are able to pass genes encoding for antibiotic resistance, such as tetracycline used to treat periodontal disease, as well as “pathogenicity islands” that allow the pathogens to evolve into different strains that can evade past treatments (B9, B16).
Do the microbes change their environment?
Metabolism
Oral bacterial metabolism is not only important for one species growth and development but important for the entire biofilm as well. For example, Streptococci spp. which is the most prevalent bacteria in the mouth ferments carbohydrates which produce lactic acid. Veillonellae in turn uses the lactic acid produced by Streptococci to promote its own growth. Studies have shown that when Streptococci are absent Veillonellae is not able to grow, so this shows that Veillonellae growth is dependent on Streptococci metabolism (B10).
Metabolism by bacteria in the mouth also produces by products that protect the mouth from incoming pathogens and acid build up. As mentioned already Veillonellae takes up lactic acid which prevents high build up of lactic acid and allowing the mouth to maintain a constant pH. S. oligogermentan has also been shown to use lactic acid and produces a by product of hydrogen peroxide. The elevated levels of hydrogen peroxide inhibit growth of A. actinomycetemcomitans which has been proven to be a big factor in periodontal disease (B2, B6).
Diseases
Dental plaque is present in both a healthy and diseased mouth, but its only when homeostasis is not maintained that problems arise. Any changes that disrupt the homeostasis of the tooth environment, such as a change in diet (endogenous/exogenous), adhesion, cofactors for growth, pH, redox potential, gases, metabolisms of other biofilm members, abrasion from swallowing, brushing and flossing, the eruption or loss of teeth, drugs, and radiation, can cause the normal microflora species to change. Diseased mouth have a greater number of gram negative cocci, rods, filaments, and anaerobic bacteria (B3, B6). These changes allow the commensal bacteria to be taken over by the pathogenic bacteria. Now the pathogenic bacteria present in the dental plaque can begin to induce changes in neighboring bacteria gene expression that will allow itself to rapidly increase its own numbers to cause diseases (B3). An example of this is when metabolic products of the bacteria lead to dental caries, better known as cavities. Today, most host’s diets contain many fermentable carbohydrates which aciduric bacteria, S. mutans and lactobacilli, can easily fermented into organic acids, such as lactic acid. The increase in acids decreases the pH on the teeth. Once the pH has dropped below 5.5 the enamel begins to demineralize. The host’s system tries to counter the demineralization of the tooth surface by having the supersaturated saliva as a buffer system. The saliva is full of calcium and phosphate that is used to re-mineralize minor chips. However, if carbon sources are depleted and the saliva becomes unsaturated, the pH cannot readjust and neutralize leading to tooth decay (B18, B3).
Changes in the supragingival plaque have been shown to influence the subgingival plaque as well. The biofilms in both the supra- and sub-gingival areas have many of the same bacteria with similar interactions. Many of the Streptococci spp., Actionmyces spp.,Prevotella spp., Propionibacterium spp., Veilonella sppl, and F. nucleatum are found in both plaque formations (B11). According to recent studies, findings have shown that with frequent professional and personal cleaning of the supragingival plaque, there is an efficacy effect on the subgingival plaque and can help reduce a person’s susceptible for diseased periodontal crevices (B17)
Gingiva
Where located?
Physical Conditions?
What are the conditions in your niche? Temperature, pressure, pH, moisture, etc.
Influence by Adjacent Communities (if any)
Is your niche close to another niche or influenced by another community of organisms?
Conditions under which the environment changes
Do any of the physical conditions change? Are there chemicals, other organisms, nutrients, etc. that might change the community of your niche.
Who lives there?
Which microbes are present?
You may refer to organisms by genus or by genus and species, depending upon how detailed the your information might be. If there is already a microbewiki page describing that organism, make a link to it.
Are there any other non-microbes present?
Plants? Animals? Fungi? etc.
Do the microbes that are present interact with each other?
Describe any negative (competition) or positive (symbiosis) behavior
Do the microbes change their environment?
Do they alter pH, attach to surfaces, secrete anything, etc. etc.
Do the microbes carry out any metabolism that affects their environment?
Do they ferment sugars to produce acid, break down large molecules, fix nitrogen, etc. etc.
Tongue
Where located?
Physical Conditions?
What are the conditions in your niche? Temperature, pressure, pH, moisture, etc.
Influence by Adjacent Communities (if any)
Is your niche close to another niche or influenced by another community of organisms?
Conditions under which the environment changes
Do any of the physical conditions change? Are there chemicals, other organisms, nutrients, etc. that might change the community of your niche.
Who lives there?
Which microbes are present?
You may refer to organisms by genus or by genus and species, depending upon how detailed the your information might be. If there is already a microbewiki page describing that organism, make a link to it.
Are there any other non-microbes present?
Plants? Animals? Fungi? etc.
Do the microbes that are present interact with each other?
Describe any negative (competition) or positive (symbiosis) behavior
Do the microbes change their environment?
Do they alter pH, attach to surfaces, secrete anything, etc. etc.
Do the microbes carry out any metabolism that affects their environment?
Do they ferment sugars to produce acid, break down large molecules, fix nitrogen, etc. etc.
Throat
Where located?
Physical Conditions?
What are the conditions in your niche? Temperature, pressure, pH, moisture, etc.
Influence by Adjacent Communities (if any)
Is your niche close to another niche or influenced by another community of organisms?
Conditions under which the environment changes
Do any of the physical conditions change? Are there chemicals, other organisms, nutrients, etc. that might change the community of your niche.
Who lives there?
Which microbes are present?
You may refer to organisms by genus or by genus and species, depending upon how detailed the your information might be. If there is already a microbewiki page describing that organism, make a link to it.
Are there any other non-microbes present?
Plants? Animals? Fungi? etc.
Do the microbes that are present interact with each other?
Describe any negative (competition) or positive (symbiosis) behavior
Do the microbes change their environment?
Do they alter pH, attach to surfaces, secrete anything, etc. etc.
Do the microbes carry out any metabolism that affects their environment?
Do they ferment sugars to produce acid, break down large molecules, fix nitrogen, etc. etc.
Current Research
1. Correlation between periodontitis and coronary heart disease
Several theories exist to explain the relationship between coronary heart disease and periodontitis. One such theory includes the role of fibrinogen in linking the two. Fibrinogen is a plasma glycoprotein synthesized in the liver. After coagulation, it is converted in Fibrin, the protein involved in blood clots. The study took 95 subjects and separated them into a healthy control group, moderately and severely chronic periodontitis group, coronary heart disease group, and moderately and severely chronic periodontitis coexisting with coronary heart disease group. After measuring routinely diagnosis procedure for both periodontitis and cardiovascular heart disease along with levels of fibrinogen, results showed fibrinogen levels in subjects with MSP and MSP+CHD to be much higher than those of the healthy controlled group. It was concluded that people with periodontitis may be more at risk for developing cardiac heart disease when taking into consideration fibrinogen as the biological basis.
2. Bacterial markers of periodontal diseases
Among the various bacteria present in the subgingival deposit, three have been the center of study due to their prevalence in periodontal disease. First, the Tannerella forsythensis, an anaerobic Gram-negative known to possess pathogenic potential. Second, the Actinobacillus actinomycetemcomitans with the ability to attach itself to the enamel surface after the consumption of sugar to produce an acid resulting in the erosion of the enamel surface. And the third, the Porphyronmonas gingivalis, a gram negative bacteria with a virulence factor allowing it to invade the gum tissue. They measure the bacteria on 495 subjects aged 6-82 with the use of the 16S ribosomal RNA based polymerase chain reaction (PCR). Their studies showed an increase in the amount of T. forsythensis and P. gingivalis on older subjects when compared to the adolescents. Furthermore, an increase in T. forsythensis was also seen within the tobacco smokers or the subjects. It was finally concluded that the T. forsythensia had a correlation between periodontal disease in tobacco smoking and elder individuals.
3. Saliva used as Biomarkers for early disease detection
Current research is being done in the mouth to help detect diseases throughout the entire body. As of now, some hard hitting diseases: cancer, cardiovascular diseases, metabolic and neurological deficits are tough to diagnose especially early on. The research is aimed at providing scientists and doctors a simple, inexpensive way to diagnose these diseases through finding disease-associated proteins and genetic markers in saliva. Saliva dectection is already being used to detect HIV, hepatitis A, B, C, and is used to monitor some drugs like marijuana. The NIDCR is currently working on finding the body’s disease-associated salivary biomarkers for those hard to diagnose diseases than developing an accurate, easy to use diagnostic method (Wong).
4. Fluorinated food
For a while now some researchers have been looking into fluorinating food sources. This is done with the thought that because fluoride is the best way of preventing big plaque build up on the tooth, since plaque is linked with gingivitis, periodontal disease, and other oral disease. This current research finds that fluorinating food in populations where fluoride toothpaste and daily brushing has almost no additional beneficial effects. The big reason in wanting to fluorinate food is too possibly fight oral diseases in other countries in which brushing is not practiced regularly. This experiment shows that without the brushing, fluorinated foods have a small but significant effect on controlling plaque it also shows that fluorinated foods should not be substituted for brushing with fluorinated toothpaste (Meyer-Luekel).
References
Chukhlovin AB, Solovyova AM, Matelo SK, Kobiyasova IV, Morosova EB, Hokhlacheva AV, Teplyakov BG, Syssoev KA, Konstantinova VE, Matelo LN, Totolian AA. “Bacterial markers of periodontal diseases and their practical significance in dentistry.” Bull Exp Biol Med. 2007 Oct;144(4):546-50. PMID: 18642710 http://www.ncbi.nlm.nih.gov/pubmed/18642710?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
Ge S, Wu YF, Liu TJ, Meng S, Zhao L. “Study of the correlation between moderately and severely chronic periodontitis and coronary heart disease”, Hua Xi Kou Qiang Yi Xue Za Zhi. 2008 Jun;26(3):262-6. PMID: 18705507 http://www.ncbi.nlm.nih.gov/pubmed/18705507?ordinalpos=18&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pub
Mydel P, Takahashi Y, Yumoto H, Sztukowska M, Kubica M, Gibson FC 3rd, Kurtz DM Jr, Travis J, Collins LV, Nguyen KA, Genco CA, Potempa J. “Roles of the host oxidative immune response and bacterial antioxidant rubrerythrin during Porphyromonas gingivalis infection.” PLoS Pathog. 2006 Jul;2(7):e76. PMID: 16895445 http://www.ncbi.nlm.nih.gov/pubmed/16895445?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
Rautemaa R, Järvensivu A, Kari K, Wahlgren J, DeCarlo A, Richardson M, Sorsa T. Oral Dis. 2004 Sep;10(5):298-305. “Intracellular localization of Porphyromonas gingivalis thiol proteinase in periodontal tissues of chronic periodontitis patients.” PMID: 15315648 http://www.ncbi.nlm.nih.gov/pubmed/15315648?ordinalpos=8&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
Yang R, Zou J, Li JY. “Study of the relationship between oral Actinomyces and childhood caries.” Hua Xi Kou Qiang Yi Xue Za Zhi. 2007 Dec;25(6):568-70. PMID: 18306628 http://www.ncbi.nlm.nih.gov/pubmed/18306628?ordinalpos=13&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
Frederick JF, Rogers EA, Marconi RT. Analysis of a growth phase regulated-two component regulatory system in the periodontal pathogen, Treponema denticola. J Bacteriol. 2008 Jul 11. PMID: 18621891 http://www.ncbi.nlm.nih.gov/pubmed/18621891?ordinalpos=3&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_RVDocSum
A S McDermid, A S McKee, and P D Marsh. “Effect of environmental pH on enzyme activity and growth of Bacteroides gingivalis W50.” Infect Immun. 1988 May; 56(5): 1096–1100. PMCID: PMC259768 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=259768
Edited by Mason Chen, Diana Kirchmann, Alvin Kwong, Lydi Martinez, Mei Ng, Gabriel Tran, Kristen Watanabe, Kathryn Yee students of Rachel Larsen