Nanotubes facilitate intercellular signaling in eukaryotic and prokaryotic cells
Overview
<http://www.sciencedirect.com/science?_ob=MiamiCaptionURL&_method=retrieve&_eid=1-s2.0-S009286741100016X&_image=1-s2.0-S009286741100016X-figs5.jpg&_cid=272196&_explode=defaultEXP_LIST&_idxType=defaultREF_WORK_INDEX_TYPE&_alpha=defaultALPHA&_ba=&_rdoc=1&_fmt=FULL&_issn=00928674&_pii=S009286741100016X&md5=b2d22cbeb0a19edbc6d02597fb061a4d>.
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki. The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Electron micrograph of the Ebola Zaire virus. This was the first photo ever taken of the virus, on 10/13/1976. By Dr. F.A. Murphy, now at U.C. Davis, then at the CDC.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
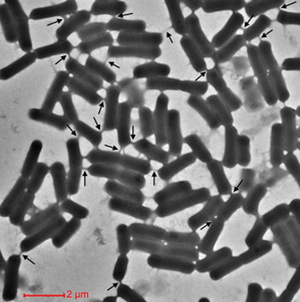
Cells encourage diversity by using several different molecular signaling mechanisms to transfer information, nutrients, organelles, or parasites. Many of these processes require direct cellular interaction for signals to be exchanged. Prokaryotes use conjugation as a means for horizontal gene transfer from one cell to another. Eukaryotes use gap junctions to permit passage of ions and small molecules between the cytoplasm of one cell to another. Many quorum sensing eukaryotes and prokaryotes use extracellular signaling molecules to synchronize their gene expression, behavior, and population density and thus exhibit multicellular properties.[1],[2] Other single-celled or multicellular organisms (and even cancerous cells) release extracellular membrane vesicles or exosomes to shuttle microRNAs, mRNAs, DNA fragments, and proteins to a recipient cell.[3] Additionally, recent research has shown nanotubes (also called tunneling nanotubes (TNTs) or membrane nanotubes (MNTs)) to transiently form between two or more eukaryotic or prokaryotic cells (Figure 1). These nanotubular channels are cell-to-cell plasma membrane connections of varying lengths that allow for bridging cargo exchange and, in some cases, multicellular properties.
Emerging research within the past 10 years has affirmed cellular nanotubes to be involved in complex signaling processes such as viral and prion pathogenesis among eukaryotic cells and metabolic or genetic exchanges among many types of bacteria. Surprisingly, the retrovirus HIV-1 has been shown to move between T-cells, macrophages, and B-cells by use of a nanotube-delivery mechanism for viral proteins.[15],[16],[20],[22] The leukemia-causing HTLV-1 similarly spreads throughout human T-cells by a similar mechanism to HIV-1, and this mechanism allows for a powerful and rapid transmission of the virus throughout the immune system. Prions (PrPSc) that cause degenerative neural disease have been shown to take over nanotubes in bone-marrow derived dendritic cells and spread by them to neural cells, allowing for a safe intercellular route to the brain.[12] Pathogens can indeed utilize already-existing nanotubes between cells, but they have been shown in some cases to even induce nanotube formation.[22] Elucidation of these eukaryotic mechanisms could provide potential targets for drug therapy in otherwise untreatable or hard-to-treat diseases. However, transport of cytoplasmic material through nanotubes is not an activity limited only to eukaryotic cells. Both similar and diverse species of bacteria have been observed to exchange not only cytoplasmic constituents, but also genetic material in the form of plasmids.[19] Dynamic nanotubes have been witnessed between the likes of B. subtilis, E. coli, Staphylococcus aureus, and Acinetobacter baylyi.[18],[19] The new discovery of a stress-induced mutualistic nutrient exchange between distantly related species of bacteria pushes the boundaries of our definition of “unicellular” organisms and prompts us to reevaluate bacteria as a multicellular, interconnected domain of life.[18]
Structure
Cellular nanotubes are membranous, tubular protrusions ranging anywhere from 50 nm to several hundred nm in diameter and, in some cases, extending hundreds of μm (several cell diameters) in length.[4],[5] First discovered by Rustom et al. 2004, these cylindrical channels were proposed to facilitate the traffic of intracellular vesicles and mediate membrane continuity between rat pheochromocytoma (PC12) cells.[5] In the same year, Önfelt et al. 2004 discovered nanotubes to transiently connect networks of immune cells.[6] As such, the mechanism for nanotube formation in eukaryotic animal cells has been largely characterized.
In human monocyte-derived macrophages, the cytoskeleton protein F-actin has been shown to support nanotubes of varying structure and function. These nanotubes also were shown to accommodate extracellular pathogens, such as the bacteria Mycobacterium bovis bacillus calmette-Guerin (BCG) that “surfed” along the surface of the nanotube and were subsequently phagocytosed by the macrophage.[7] Bridging between white blood cells, thin (<0.7 μm in diameter) nanotubes are composed of F-actin and allow for unidirectional movement of intracellular cargo within the nanotube.[7] Contrarily, wider (>0.7 μm in diameter) nanotubes composed of both actin and microtubules were found to aid in the bidirectional movement of organelles (lysosomes and mitochondria) and lipid vesicles between the macrophages [7] This study by Önfelt et al. 2004 suggested a defensive functionality of nanotubes, which could bind extracellular bacteria and shuttle them toward the macrophage for phagocytosis.
Although F-actin was generally accepted to be a necessary component of eukaryotic nanotubes, Veranič et al. 2008 characterized novel type II nanotubes between RT4 and T24 urothelial cells that were long and stable due to a presence of cytokeratin and absence of F-actin. These type II nanotubes, which could extend up to several 100 μm in length, seemed to lack any signaling functionality but could connect cells that were in the process of dissociating for more than 2 hours. [9] Likewise, Wang et al. 2012 found nanotubes between neurons astrocytes that instead all contained microtubules—these nanotubes were actually longer if they lacked F-actin than if they were supported by it. [8]
Formation
<http://link.springer.com/article/10.1007%2Fs11427-013-4548-3>.
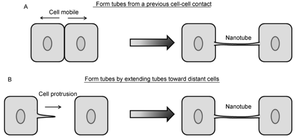
Contrarily, nanotubes are inhibited by chemical fixation, mechanical stress, and prolonged light exposure, making these structures intricately difficult to study.[28] However, through different methods, nanotubes have been found to form in two distinct general ways that differ per animal cell type (Figure 2).[10],[11] One possibility is that actin polymerizes within a cell, causing a protrusion that can contact another cell. Such an approach would be useful for immobile or less-mobile cells like rat pheochromocytoma PC12.[10], [23] Alternatively, cells that are in prolonged contact for a several minutes could move apart, trailing a newly formed nanotube between them in the separated space. This strategy is used by mobile cell types, such as T-cells that have an increased chance for collision with other cells.[10],[15]
A diverse range of conditions has been found to contribute to nanotube formation in animal cells.[25] Blood serum starvation and Lipopolysaccharide-induced inflammation has been shown to stimulate formation of nanotubes up to 330 μm long in mice cornea.[26] Oxidative cellular stress in the form of hydrogen peroxide to induce nanotubes between neural cells.[21] The interaction of the Fas ligand with its receptor and subsequent activation of Rho GTPase has also been shown to cause nanotube formation in T-cells; this mechanism is surprising, as the stimulation of the Fas ligand has been previously characterized to induce apoptosis.[25],[27] Interestingly, viral infection has been shown to induce nanotube formation. Although HIV-1 does not cause increased amounts of nanotubes on or between T-cells, it correlated with an increased number of nanotubes on human macrophages. Following a similar trend but with a different cell type, the retrovirus HTLV was found to stimulate a larger number of nanotubes on unactivated, static T-cells.[15],[22]
HIV Pathogenesis
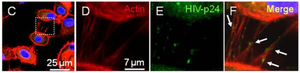
<http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2701345/figure/F5/>.
Include some current research in each topic, with at least one figure showing data.
HTLV Pathogenesis
Overall paper length should be 3,000 words, with at least 3 figures.
Prion Pathogenesis

Include some current research in each topic, with at least one figure showing data.
Prion Pathogenesis
Include some current research in each topic, with at least one figure showing data.
Genetic and cytoplasmic exchange in B. subtilis
Include some current research in each topic, with at least one figure showing data.
Metabolic mutualism between E. coli and Acinetobacter baylyi
Include some current research in each topic, with at least one figure showing data.
References
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
