Nocardia: Difference between revisions
Acjenkins2 (talk | contribs) |
Acjenkins2 (talk | contribs) |
||
| Line 37: | Line 37: | ||
[3] Stothard P, Van Domselaar G, Shrivastava S, Guo A, O'Neill B, Cruz J, Ellison M, Wishart DS (2005) BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res 33:D317-D320 | [3] Stothard P, Van Domselaar G, Shrivastava S, Guo A, O'Neill B, Cruz J, Ellison M, Wishart DS (2005) BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res 33:D317-D320 | ||
[4] Munoz J, Mirelis B, et al., Clinical and microbiological features of nocardiosis 1997-2003. | |||
=Figures= | =Figures= | ||
Revision as of 01:04, 13 May 2015
Classification
Domain: Bacteria Phylum: Actinobacteria Class: Actinobacteria Order: Actinomycetales Family: Nocardiaceae Genus: Nocardia Species: N. asteroides, N. brasiliensis, N. farcinica, N. veterana
Description
The genus Nocardia was first discovered by Edmund Nocard, a French veterinarian, in 1889, as the main cause of bovine infections []. The species of Nocardia genus are gram-positive and rod-shaped bacteria. They are reliant on getting their nourishment from dead and/or decaying organic material, making these bacteria saprophytic. Nocardia are commonly found in soils, plants, animal tissues, and human tissues [1].
The bacteria in this species have been common to diseases. Nocardiosis, the most common disease associated with these genus, which is an infection disease affecting either the lungs or the entire body. Also, the inhalation of Nocardia species can lead to pneumonia, as well as skin infections if they enter through open wounds [2].
Ecology and Significance
Genome Structure
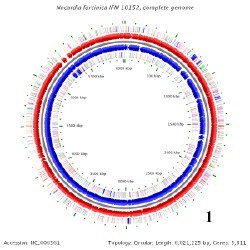
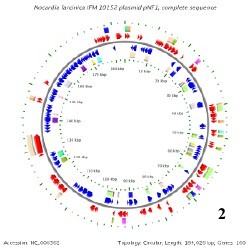
There is only one known genome in the Nocardia species and that belongs to Nocardia farcinica. This genome contains a circular chromosome that is 6,021,225 base pairs long, containing 5,811 genes, and having a GC content around 71%. N. farcinica has two plasmids that go along with its chromosome, pNF1 and pNF2. pNF1 is 184,027 base pairs long, consisting of 160 genes, and with a GC content of 67%. pNF2 is 87,093 base pairs long, made up of 93 genes, and has a GC content of 60%. All Of these information pertaining to the genome have been determined from 16S rRNA sequencing. [3]
Below is BacMap Genome Mapping, courtesy of Stothard P, Van Domselaar G, Shrivastava S, Guo A, O'Neill B, Cruz J, Ellison M, Wishart DS (2005) BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res 33:D317-D320.
 ****
**** ****
****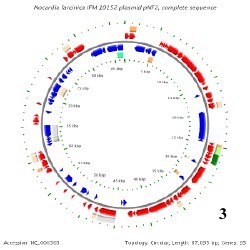
Image 1: N. farcinica Chromosome
Image 2: pNF1 (Plasmid 1)
Image 3: pNF2 (Plasmid 2)
Metabolism
References
[1] Jun Ishikawa, Atsushi Yamashita, Yuzuru Mikami, Yasutaka Hoshino, Haruyo Kurita, Kunimoto Hotta, Tadayoshi Shiba, and Masahira Hattori. The complete genomic sequence of Nocardia farcinica IFM 10152. 101:41 (2004): Web.
[2] Segawa, Shunsuke. "Identification of Nocardia species using matrix-assisted laser desorption/ionization-time-of-flight-mass spectrometry." 12:6 (2015): Web.
[3] Stothard P, Van Domselaar G, Shrivastava S, Guo A, O'Neill B, Cruz J, Ellison M, Wishart DS (2005) BacMap: an interactive picture atlas of annotated bacterial genomes. Nucleic Acids Res 33:D317-D320
[4] Munoz J, Mirelis B, et al., Clinical and microbiological features of nocardiosis 1997-2003.
Figures
[1F]
[4] [2F] [5] [3F] [6] [4F] [7] [5F] [Original Figure. Author: Pawan Dhaliwal] [6F]
http://microbewiki.kenyon.edu/index.php/File:Lorenzo.gif
[7F] [8]
Author
Page authored by Alex Jenkins, student of Prof. Katherine Mcmahon at University of Wisconsin - Madison.
