Ochrobactrum anthropi: Difference between revisions
| Line 18: | Line 18: | ||
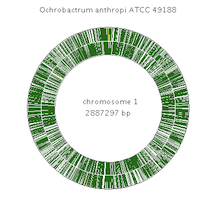
The 4.8-Mb genome of ''O. anthropi'' consists of two identical circular chromosomes. Both Chromosomes have an average C+G content of 56% and together comprise 4,424 protein-coding genes (~87% coding), along with 31 pseudogenes and 73 structural RNA's (rRna, tRNA, and small RNA).1 | The 4.8-Mb genome of ''O. anthropi'' consists of two identical circular chromosomes. Both Chromosomes have an average C+G content of 56% and together comprise 4,424 protein-coding genes (~87% coding), along with 31 pseudogenes and 73 structural RNA's (rRna, tRNA, and small RNA).1 | ||
The O. anthropi genome also contains four plasmids. pOAN01, pOAN02, and pOAN03 have the expected characteristics of alphaproteobacterial plasmids, but pOAN04 doesn't - It lacks known replication, partition and conjugative systems.1 | The O. anthropi genome also contains four plasmids. pOAN01, pOAN02, and pOAN03 have the expected characteristics of alphaproteobacterial plasmids, but pOAN04 doesn't - It lacks known replication, partition and conjugative systems.1 | ||
[[Image:Chromosome1.png|frame|right| | [[Image:Chromosome1.png|frame|right|100px|Genome of O. anthropi]] | ||
==Structure and Metabolism== | ==Structure and Metabolism== | ||
Revision as of 03:24, 2 May 2012
A Microbial Biorealm page on the genus Ochrobactrum anthropi
Classification
Higher order taxa
Bacteria; Proteobacteria; Alphaproteobacteria; Rhizobiales; Brucellaceae; Ochrobacterium; Ochrobacterium anthropi
Species
Description and significance
O. anthropi,formerly known as Achromobacter species (CDC group Vd), is a common soil alphaproteobacteria that colonizes a wide spectrum of organisms and is being increasingly recognized as a potentially problematic opportunistic and nosocomial human pathogen.1 Though it is extremely rare for it to affect a healthy human, there have been numerous published cases being associated with the presence of indwelling medical devices, such as central venous catheters, drainage tubes, and intraperitoneal catheters.2 It is also characterized by a broad spectrum of antibiotic resistance.2
Genome structure
The 4.8-Mb genome of O. anthropi consists of two identical circular chromosomes. Both Chromosomes have an average C+G content of 56% and together comprise 4,424 protein-coding genes (~87% coding), along with 31 pseudogenes and 73 structural RNA's (rRna, tRNA, and small RNA).1 The O. anthropi genome also contains four plasmids. pOAN01, pOAN02, and pOAN03 have the expected characteristics of alphaproteobacterial plasmids, but pOAN04 doesn't - It lacks known replication, partition and conjugative systems.1
Structure and Metabolism
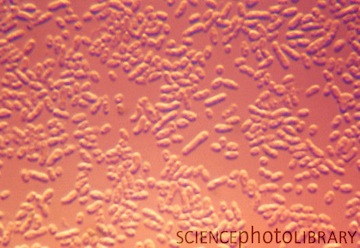
O. anthropi strains are rod shaped, non-pigmented, and motile by means of peritrichous flagella.6 They are strictly aerobic, gram negative bacilli that produce oxidase, urease, and acid from several carbohydrates (glucose, fructose, arabinose, xylose, mannitol, inositol, and ethanol). They also reduce both nitrate and nitrite by assimilation.3,6 After growth on nutrient agar for 24 hours, the colonies are typically circular, low convex, about 1 mm in diameter, smooth, shining, and entire.
Ecology
O. anthropi is a versatile alphaproteobacterium with ability to colonize an exceptionally wide variety of habitats, from hostile environments such as polluted soil, to water, plants, nematodes, insects, animals, and man.5 According to their habitat and/or to the relationships with their host, structure is varied.5
Pathology
Although cases of O. anthropi being pathogenic are rare, there are several of documented cases showing that it is possible for individuals with an underlying medical condition. Among these individuals, those with an indwelling medical device, such as catheters and drainage tubes, are most susceptible. This is likely due to its ability to adhere to various synthetic materials.3 The effects of O. anthropi when pathogenic seem to vary greatly depending on the host. Some reported cases include life-threatening infections, such as endocarditis, whereas others have completely different effects.4 O. anthropi has a very broad spectrum of antibiotic resistance which makes it is difficult to treat.2
References
List your references here with hyperlinks to the papers or websites when possible. Also, provide the DOI number for articles. For example:
1. Patrick S. G. Chain, Dorothy M. Lang, Diego J. Comerci, Stephanie A. Malfatti, Lisa M. Vergez, Maria Shin, Rodolfo A. Ugalde, Emilio Garcia, Marcello E. Tolmasky. Genome of Ochrobactrum anthropi ATCC 49288T a Versatile Opportunistic Pathogen and Symbiont od Several Eukaryotic Hosts. Journal of Bacteriology 2011.
2. Adrian Kettaneh, Francois-Xavior Weill, Isabelle Poilane, Oliver Fran, Michel Thomas, Jean-Louis Herrmann, Laurent Hocqueloux. Septic Shock Caused by Ochrobactrum anthropi in an Otherwise Healthy Host. J. Clin Microbiol 2003
3. Alnor D, Frimodt-Moller N, Espersen F, Frederiksen W. Infections with the Unusual Human Pathogens Agrobacterium Species and Ochrobactrum anthropi. Clinical Infectious Diseases 1994
4. Mahmood MS, Sarwari AR, Khan MA, Sophie Z, Khan E, Sami S. Infective Endocarditis and Septic Embolization with Ochrobactrum anthropi: case report and review of literature. The Journal of Infection 2000.
5. Sara Romano, Fabian Aujoulat, Estelle Jumas-Bilak, Anges Masnou, Jean-Luc Jeannot, Envold Falsen, Helene Marchandin, Corinne Teyssier. Multilocus Sequence Typing Supports the Hypothesis that Ochrobactrum anthropi Displays a Human-Associated Subpopulation. BMC Microbiology 2009
6. B. Holmes, M. Popoff, M. Kiredjian, M. Kersters. Ochrobactrum anthropi gen. nov., sp. nov. from Human Clinical Specimens and Previously Known as Group Vd. International Journal of Systematic and Evolutionary Microbiology 2012.
Edited by Eric Beaver of Dr. Lisa R. Moore, University of Southern Maine, Department of Biological Sciences, http://www.usm.maine.edu/bio