Oenococcus oeni: Difference between revisions
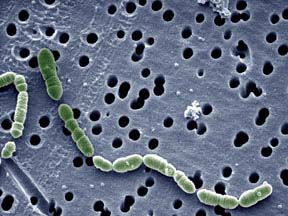
(Created page with "ated}} thumb|1000px|FIGURE 1. Typical shape and morphology of Oenococcus oeni (7). ==Classification== Domain: Bacteria Phylum: ''Firmicutes'' Cla...") |
No edit summary |
||
| Line 1: | Line 1: | ||
{{Uncurated}} | |||
[[Image:oenococcus.jpg|thumb|1000px|FIGURE 1. Typical shape and morphology of Oenococcus oeni (7).]] | [[Image:oenococcus.jpg|thumb|1000px|FIGURE 1. Typical shape and morphology of Oenococcus oeni (7).]] | ||
==Classification== | ==Classification== | ||
Domain: Bacteria | Domain: Bacteria | ||
Phylum: ''Firmicutes'' | Phylum: ''Firmicutes'' | ||
Class: ''Bacilli'' | Class: ''Bacilli'' | ||
Order: ''Lactobacillales'' | Order: ''Lactobacillales'' | ||
Family: ''Leuconostocaceae'' | Family: ''Leuconostocaceae'' | ||
Genus: ''Oenococcus'' | Genus: ''Oenococcus'' | ||
Species: ''oeni'' (1) | Species: ''oeni'' (1) | ||
[http://www.ncbi.nlm.nih.gov/Taxonomy/ NCBI] link to find] | [http://www.ncbi.nlm.nih.gov/Taxonomy/ NCBI] link to find] | ||
===Species=== | ===Species=== | ||
| Line 32: | Line 23: | ||
==Description and Significance== | ==Description and Significance== | ||
Oenococcus oeni (previously classified as Leuconostoc oenos) is a gram-positive, nonmotile, chemoorganotrophic lactic acid bacterium that grows in chains of circular to ellipsoidal cells (2). Unlike many bacteria, it thrives in the harsh environment of wine and is considered a beneficial to wine quality and character, rather an agent of damaging value (3). O. oeni is beneficial to wine and the entire field of oenology because of its primary ability to perform malolactic fermentation, the crucial secondary fermentation in the production of wine of converting malic acid to lactic acid. It occurs naturally in musts, wines, and fruit mashes (4). | |||
==Genome Structure== | ==Genome Structure== | ||
Any strain of O. | In 1995, Dicks et al. sought to reclassify Leuconostoc oenos as Oenococcus oeni because there is little DNA homology between the two genera (2). Oenococcus differs further from Leuconostoc from its abilities to use saccharose, lactose, and maltose as a substrate (5). In 2006, a second Oenococcus species was identified (6). | ||
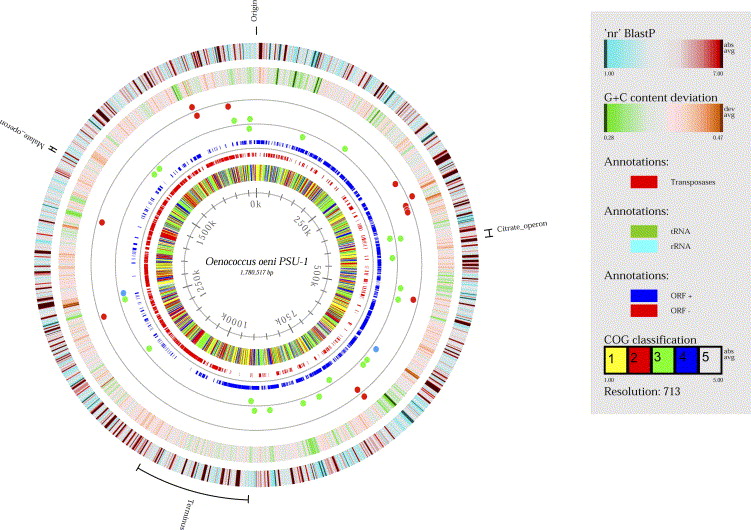
Any strain of O. oeni has a circular genome of 1780517 basepair nucleotides, 1691 protein-coding genes, and 51 RNA genes (7). | |||
[[Image:Oenococcus_genome.jpg|thumb|1000px|FIGURE 2. Genome of Oenococcus oeni PSU-1 (8).]] | |||
Describe the size and content of the genome. Other interesting features? What is known about its sequence? | |||
==Cell Structure, Metabolism and Life Cycle== | ==Cell Structure, Metabolism and Life Cycle== | ||
Oenococcus oeni is heterofermentative and can grow in acidic conditions below pH 3.0 (2). It is ethanol tolerant at 10%. The cocci are ellipsoidal to spherical in shape and usually grow in chains or pairs. Optimal growth occurs on sugar and protein rich media, like grape or tomato juice. | |||
Lactic acid bacteria, like O. oeni, are typically non-sporulating and | |||
==Ecology and Pathogenesis== | ==Ecology and Pathogenesis== | ||
Habitat; symbiosis; biogeochemical significance; contributions to environment.<br> | Habitat; symbiosis; biogeochemical significance; contributions to environment.<br> | ||
Oenococcus oeni stabilizes wine communities by consuming available nutrients and lowering potential growth of other microbes, but its malolactic fermentation can be beneficial or detrimental to the production of wine depending on grapes, climate, and style of wine. Industrial winemakers use a standardized strain of O. oeni, but the many external and environmental variables can dictate the success of the wine. | |||
==References== | ==References== | ||
(1) [http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=Retrieve&list_uids=16644 NCBI Genome Project] | |||
(2) [http://www.ncbi.nlm.nih.gov/pubmed/7537074 Dicks, L. M. T., Dellaglio, F., Collins, M. D. “Proposal To Reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov”. “International Journal of Systematic Bacteriology”. 1995. Volume 445. P. 395-397] | |||
(1) | (3) [http://www.biomedcentral.com/1471-2164/13/373 Borneman, A. R., McCarthy, J. M., Chambers, P. J., Bartowsky, E. J. “Comparative analysis of the Oenococcus oeni pan genome reveals genetic diversity in industrially-relevant pathways”. 2012. BMC Genomics. Volume 13. P. 373] | ||
(2) [Dicks, L. M. T., Dellaglio, F., Collins, M. D. “Proposal To Reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov”. “International Journal of Systematic Bacteriology”. 1995. Volume 445. P. 395-397] | (4) [http://genome.jgi-psf.org/oenoe/oenoe.home.html Oenococcus oeni PSU-1. The Regents of the University of California] | ||
(3) [http://www.biomedcentral.com/1471-2164/13/373 Borneman, A. R., McCarthy, J. M., Chambers, P. J., Bartowsky, E. J. “Comparative analysis of the Oenococcus oeni pan genome reveals genetic diversity in industrially-relevant pathways”.] | (5) [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2009.04428.x/pdf Solieri, L., Genova, F., De Palola, M., Giudici, P. “Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures”. “Journal of Applied Microbiology”. 2010. Volume 108. P. 285-298] | ||
(4) [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2672.2009.04428.x/pdf Solieri, L., Genova, F., De Palola, M., Giudici, P. “Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures”. “Journal of Applied Microbiology”. 2010. Volume 108. P. 285-298] | (6) [http://ijs.sgmjournals.org/content/56/10/2345.long Endo, A., Okada, S. “Oenococcus kitaharae sp. nov., a non-acidophilic and non-malolactic-fermenting oenococcus isolated from a composting distilled shochu residue”. “International Journal of Systematic and Evolutionary Microbiology”. 2006. Volume 56. P. 2345-2348] | ||
(7) [http://www.ncbi.nlm.nih.gov/pubmed/17030793 Kakarova, K., et al. “Comparative genomics of the lactic acid bacteria” . “Proc. Natl. Acad. Sci. USA.”. 2006. Volume 103. P. 15611-6] | |||
(8) [http://www.sciencedirect.com/science/article/pii/S0168644505000355 Mills, D. A., Rawsthorne, H., Parker, C., Tamir, D., Makarova, K. “Genomic analysis of Oenococcus oeni PSU-1 and its relevance to winemaking”. “FEMS Microbiology Reviews”. 2005. Volume 29. P. 465-475] | |||
==Author== | ==Author== | ||
Page authored by Drew Wehrle student of Prof. Jay Lennon at Indiana University. | Page authored by Drew Wehrle student of Prof. Jay Lennon at Indiana University. | ||
<!-- Do not remove this line-->[[Category:Pages edited by students of Jay Lennon at Indiana University]] | <!-- Do not remove this line-->[[Category:Pages edited by students of Jay Lennon at Indiana University]] | ||
. | |||
Revision as of 15:56, 28 April 2014
Classification
Domain: Bacteria Phylum: Firmicutes Class: Bacilli Order: Lactobacillales Family: Leuconostocaceae Genus: Oenococcus Species: oeni (1)
NCBI link to find]
Species
|
NCBI: Taxonomy |
Oenococcus oeni
Description and Significance
Oenococcus oeni (previously classified as Leuconostoc oenos) is a gram-positive, nonmotile, chemoorganotrophic lactic acid bacterium that grows in chains of circular to ellipsoidal cells (2). Unlike many bacteria, it thrives in the harsh environment of wine and is considered a beneficial to wine quality and character, rather an agent of damaging value (3). O. oeni is beneficial to wine and the entire field of oenology because of its primary ability to perform malolactic fermentation, the crucial secondary fermentation in the production of wine of converting malic acid to lactic acid. It occurs naturally in musts, wines, and fruit mashes (4).
Genome Structure
In 1995, Dicks et al. sought to reclassify Leuconostoc oenos as Oenococcus oeni because there is little DNA homology between the two genera (2). Oenococcus differs further from Leuconostoc from its abilities to use saccharose, lactose, and maltose as a substrate (5). In 2006, a second Oenococcus species was identified (6). Any strain of O. oeni has a circular genome of 1780517 basepair nucleotides, 1691 protein-coding genes, and 51 RNA genes (7).
Describe the size and content of the genome. Other interesting features? What is known about its sequence?
Cell Structure, Metabolism and Life Cycle
Oenococcus oeni is heterofermentative and can grow in acidic conditions below pH 3.0 (2). It is ethanol tolerant at 10%. The cocci are ellipsoidal to spherical in shape and usually grow in chains or pairs. Optimal growth occurs on sugar and protein rich media, like grape or tomato juice. Lactic acid bacteria, like O. oeni, are typically non-sporulating and
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
Oenococcus oeni stabilizes wine communities by consuming available nutrients and lowering potential growth of other microbes, but its malolactic fermentation can be beneficial or detrimental to the production of wine depending on grapes, climate, and style of wine. Industrial winemakers use a standardized strain of O. oeni, but the many external and environmental variables can dictate the success of the wine.
References
(1) NCBI Genome Project (2) Dicks, L. M. T., Dellaglio, F., Collins, M. D. “Proposal To Reclassify Leuconostoc oenos as Oenococcus oeni [corrig. gen. nov., comb. nov”. “International Journal of Systematic Bacteriology”. 1995. Volume 445. P. 395-397] (3) Borneman, A. R., McCarthy, J. M., Chambers, P. J., Bartowsky, E. J. “Comparative analysis of the Oenococcus oeni pan genome reveals genetic diversity in industrially-relevant pathways”. 2012. BMC Genomics. Volume 13. P. 373 (4) Oenococcus oeni PSU-1. The Regents of the University of California (5) Solieri, L., Genova, F., De Palola, M., Giudici, P. “Characterization and technological properties of Oenococcus oeni strains from wine spontaneous malolactic fermentations: a framework for selection of new starter cultures”. “Journal of Applied Microbiology”. 2010. Volume 108. P. 285-298 (6) Endo, A., Okada, S. “Oenococcus kitaharae sp. nov., a non-acidophilic and non-malolactic-fermenting oenococcus isolated from a composting distilled shochu residue”. “International Journal of Systematic and Evolutionary Microbiology”. 2006. Volume 56. P. 2345-2348 (7) Kakarova, K., et al. “Comparative genomics of the lactic acid bacteria” . “Proc. Natl. Acad. Sci. USA.”. 2006. Volume 103. P. 15611-6 (8) Mills, D. A., Rawsthorne, H., Parker, C., Tamir, D., Makarova, K. “Genomic analysis of Oenococcus oeni PSU-1 and its relevance to winemaking”. “FEMS Microbiology Reviews”. 2005. Volume 29. P. 465-475
Author
Page authored by Drew Wehrle student of Prof. Jay Lennon at Indiana University. .