Paenibacillus polymyxa
Note: Still in the process of being edited.
Classification
Domain (Bacteria); Phylum (Firmicutes); Class (Bacilli); Order (Bacillales); Family (Paenibacillaceae); Genus (Paenibacillus)
Species
Paenibacillus polymyxa
*Formerly known as Bacillus polymyxa
Discription and Significance
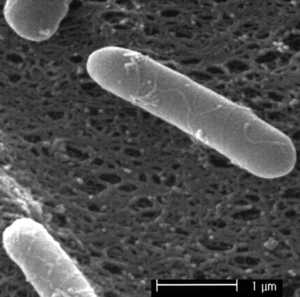
Paenibacillus polymyxa is a non-pathogenic and endospore-forming bacterium which is found in environments such as soil near plant roots and marine sediment (Timmusk)(Ravi). P. polymyxa is a Gram-positive, rod-shaped bacterium (Zengguo et al., 2007). It is also a motile bacterium that has peritrichous flagella for motion (Zengguo). The wide range of capabilities of this bacteria are to fix nitrogen, produce hormones that promote plant growth, produce hydrolytic enzymes, to produce antibiotics against harmful plant and human microorganisms (Lal). It can also help plant in absorption of phosphorus and enhance soil porosity (Timmusk 1999). Such potentials these microbes a great role in ecosystem functioning and industrial process (Lal 2010).
In agricultural ecosystems P. polymyxa can promote plant growth through three mechanisms. The first mechanism is production of hormones like cytokinins, auxins, ethylene and gibberellins (Timmusk 1999). These compounds increase root expansion and plant growth. The second mechanism is production of antibiotics and promoting immunity of rhizosphere. Heulin et al. (1994) observed that antagonistic activity of P. polymyxa decreased the activity of two plant pathogenic fungus (Gaeumannomyces graminis var. tritici and Fusarium oxysporum). The third mechanism is the bacterium’s nitrogen fixing ability that can promote plant absorbable N in the soil. In addition, soil fluctuation and porosity is improved due to organic compounds released from P. polymyxa to soil (Timmusk 1999).
The wide ranges of applications of P. polymyxa in industry are due to secondary metabolites produced by these bacteria. The antimicrobial compound is effective against a wide range of Gram-positive and Gram-negative bacterial species therefore the bacteria can be used in biopreservation of food and medical applications. Metabolites such as polymyxin E1 and a lantibiotic also decrease the colonization of pathogens in poultry and shrimp larvaes (Lal? 8). The other area of application of metabolites produced by the bacteria is bioflocculation caused by metabolites such as 2,3-butanediol (BDL). The bacterium has been used for separation of hematite, pyrite and chalcopyrite (source). Waste and tap water treatment and fermentation are other areas of industrial application of P. polymyxa.
Genome Structure
Paenibacillus was originally classified under the genus Bacillus until it became its own genus in 1993 (Ash, 1993). This distinction was made using comparative analysis of the 16S rRNA gene sequence of three different bacilli which showed enough phylogenetic distance from Bacillus subtilis to warrant a new genus (Ash, 1993). Paenibacillus (paene + Bacillus) means almost Bacillus in Latin.
P. polymyxa SC2 and P. polymyxa E681 have had their entire genomes sequenced. The complete genome of P. polymyxa SC2 is composed of a 5.7 Mb circular chromosome with about 5,400 coding genes and a 510 kb plasmid with about 649 coding genes (Mingchao et al., 2011). The complete genome of P. polymyxa E681 is composed of a 5.3 Mb circular chromosome with about 4,800 genes and no plasmids (Kim et al., 2010). P. polymyxa SC2 and E681 have 54.58% and 45.80% G+C content respectively (Mingchao and Kim). It was found that P. polymyxa has several genes involved in antibiotic biosynthesis encoded in the chromosome (Mingchao).
Cell Structure, Metabolism, and Life Cycle
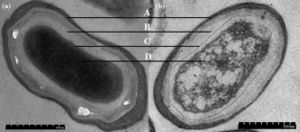
P. polymyxa is an anaerobic nitrogen-fixing Gram-positive bacterium that is rod-shaped, 0.6 by 3.0 μm in size, and produces pale colonies (Zengguo). For reproduction it differentiates into ellipsoidal spores which distinctly swell the mother cell (source). It can germinate when conditions are more suitable (Huo 2010). When P. polymyxa isn’t in an endospore form, it has peritrichous flagella that aid in motility and swarming (Zengguo).
P. polymyxa is a chemoorganoheterotroph that can fix atmospheric nitrogen and is a facultative anaerobe (Vasan). As a facultative anaerobe, it can perform aerobic respiration in the presence of oxygen or switch to fermentation when oxygen levels are low. It can use a variety of organic carbon sources such as glucose, sucrose, maltose, and arabinose and can produce a number of metabolites such as acetoin, lactate, and ethanol (de Mas). It is also a mesophile that grows optimally around 30°C and the optimum pH is around 4-7 (Vasan). P. polymyxa also has the ability to produce H2 gas as a byproduct during a fermentative process that is affected by both pH and temperature (Watanapokasin). This fermentative process, called acetogenesis, yields acetate, hydrogen, and CO2, which can be used as precursors by methanogens to produce methane (Watan).
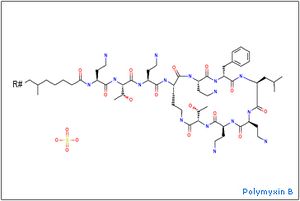
P. polymyxa has the unique capability of synthesizing antibiotics/antifungal compounds. These three of these compounds include polymxin, paenibacillin (Zengguo), and fusaricidin (Choi). Spore germination of P. polymyxa can be influenced by many factors including heat activation, fructose plus L-alanine (Huo 2010), and organic acids affect the heat resistance of spores (Casadei).
Ecology and Pathogenesis
P. polymyxa is found in variety of environments as soils, roots, rhizosphere plants and marine sediments [8]. In the rhizosphere, P. polymyxa has pathogenic traits against deleterious microorganisms (mainly fungi). It also has a symbiotic relationship with plants by invading their roots and forming biofilms (Timmusk et al, 2005). Production of hormones and nitrogen fixation are other beneficial activities of the bacteria in soil and rhizosphere. P. polymyxa also has antagonistic activity against Vibrio species and many other human and animal pathogenic microorganisms (Ravi). Therefore, it has been used in production of commercial antimicrobials (Liebminger et al., 2011).
This bacterium produces secondary metabolites that have wide applications in agricultural ecosystems, biopreservation in food and medicine industry, bioflocculation in waste water and mineral processing (Kim et al., 2010).
P. polymyxa has a unique capability of protecting tomato seedlings from bacterial wilt (Li 2011). Bacterial wilt is caused by Ralstonia solanacearum, a bacterium found in the soil that infects plants (Chavez). R. solanacearum invades the plant through the roots and colonizes in the vascular bundles in the xylem vessels (Chavez). As it grows and multiplies, it blocks the transportation of water and nutrients. P. polymyxa can prevent this bacterial wilt by colonizing and forming a biofilm around the roots of the tomato seedling, preventing the entrance of R. solanacearum.
P. polymyxa also has potential uses in bioremediation. It surrounds itself with a compound called exopolysaccharide (or extracellular polymeric substance), which is important for biofilm formation and adhering to plant roots and soil particles. This exopolysaccharide can be used as an inexpensive and easily cultivable compound to remove cadmium (Cd2+) from aqueous solutions (Mokaddem). This is achieved by the absorption of cadmium in the aqueous solution into the exopolysaccharide of P. polymyxa (Mokaddem).
Additionally, P. polymyxa can be used in the bioremediation removal of reactive blue 4 (RB4), a dye used on fabrics that is not readily removed from water by wastewater treatment processes (Watanapokasin). Watanapokasin et al. observed decolorization with dye removal by P. polymyxa along with a hydrogen byproduct that could be used as a potential energy source. This decolorization occurred via a process called acetogenesis.
References
Authors
Note: Still in the process of being edited.
Page authored by Sarah Kuenzel and Moslem Ladoni, students of Prof. Jay Lennon at Michigan State University.
