Porphyromonas gingivalis: Difference between revisions
| Line 33: | Line 33: | ||
[[Image:Klein_comparison.png|frame|right|x250px| Comparisons and overlap of essential genes and the core genome of ''P. gingivalis'' type strain ATCC 33227 to the entire curated Database of Essential Genes<ref name="Klein"/>]] | [[Image:Klein_comparison.png|frame|right|x250px| Comparisons and overlap of essential genes and the core genome of ''P. gingivalis'' type strain ATCC 33227 to the entire curated Database of Essential Genes<ref name="Klein"/>]] | ||
The whole genome sequence of ''P. givivalis'' type strain ATCC 33227 was determined in 2008 by Naito ''et al.''<ref name="Naito"/>, and deposited as Genbank accession AP009380<ref name="AP009380"/>. The genome comprises a single circular chromosome is 2.35 million bp in size, averaging 48.4% G+C content, and is absent of a plasmid. Coding sequence coverage of the chromosome is 86.1%. The 2090 protein coding sequences identified have an average length of 970 bp.<ref name="Naito"/> Mutagenesis experiments have identified 463 genes essential for ''in vitro'' viability for strain ATCC 33227, an of those, 424 are contained within the core genome of 1476 genes. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
Revision as of 05:43, 23 September 2016
Ron Ramsay, Bench C, September 23, 2016 [3]
Classification
Higher order taxa
(domain) Bacteria — (phylum) Bacteroidetes — (class) Bacteroidia — (order) Bacteroidales — (family) Porphyromonadaceae — (genus) Porphyromonas [4][5]
Species
Porphyromonas gingivalis
Type strain: 2561 = ATCC 33277 = BCRC 14417 = CCRC 14417 = CCUG 25893 = CCUG 25928 = CIP 103683 = Coykendall 2561 = DSM 20709 = JCM 12257 = KCTC 5121 = NCTC 11834 = Slots 2561 [5][6]
Description and significance
Give a general description of the species (e.g. where/when was it first discovered, where is it commonly found, has it been cultured, functional role, type of bacterium [Gram+/-], morphology, etc.) and explain why it is important to study this microorganism. Examples of citations [1], [2]
Etymology: N.L. fem. dim. n. Veillonella, named after Adrien Veillon, the French microbiologist who isolated the type species.
Genome structure
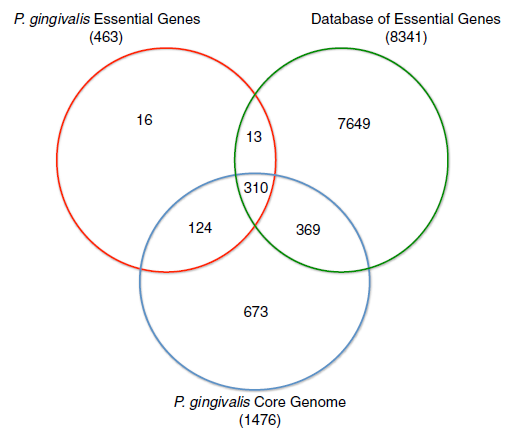
The whole genome sequence of P. givivalis type strain ATCC 33227 was determined in 2008 by Naito et al.[11], and deposited as Genbank accession AP009380[12]. The genome comprises a single circular chromosome is 2.35 million bp in size, averaging 48.4% G+C content, and is absent of a plasmid. Coding sequence coverage of the chromosome is 86.1%. The 2090 protein coding sequences identified have an average length of 970 bp.[11] Mutagenesis experiments have identified 463 genes essential for in vitro viability for strain ATCC 33227, an of those, 424 are contained within the core genome of 1476 genes.
Cell structure and metabolism
Cell wall, biofilm formation, motility, metabolic functions.
Ecology
Aerobe/anaerobe, habitat (location in the oral cavity, potential other environments) and microbe/host interactions.
Pathology
Do these microorganisms cause disease in the oral cavity or elsewhere?
Application to biotechnology
Bioengineering, biotechnologically relevant enzyme/compound production, drug targets,…
Current research
Summarise some of the most recent discoveries regarding this species.
References
- ↑ Nakayama, K. (2015). Porphyromonas Gingivalis and Related Bacteria: From Colonial Pigmentation to the Type Ix Secretion System and Gliding Motility. J Periodontal Res 50:1-8. doi:10.1111/jre.12255.
- ↑ 2.0 2.1 Shah, H. N. and M. D. Collins. (1988). Proposal for Reclassification of Bacteroides Asaccharolyticus, Bacteroides Gingivalis, and Bacteroides Endodontalis in a New Genus, Porphyromonas. Int J Syst Evol Microbiol 38:128-31. doi:10.1099/00207713-38-1-128.
- ↑ MICR3004: Microbial Genomics. University of Queensland. MICR3004 course information
- ↑ NCBI Taxonomy Browser
- ↑ 5.0 5.1 StrainInfo.net. Porphyromonas gingivalis
- ↑ LPSN: List of prokaryotic names with standing in nomenclature. Porphyromonas
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedNobody - ↑ Mysak, J., S. Podzimek, P. Sommerova, Y. Lyuya-Mi, J. Bartova, T. Janatova, J. Prochazkova, and J. Duskova. (2014). Porphyromonas Gingivalis: Major Periodontopathic Pathogen Overview. J Immunol Res 2014:8. http://dx.doi.org/10.1155/2014/476068.
- ↑ Tan, K. H., C. A. Seers, S. G. Dashper, H. L. Mitchell, J. S. Pyke, V. Meuric, N. Slakeski, S. M. Cleal, J. L. Chambers, M. J. McConville, and E. C. Reynolds. (2014). Porphyromonas Gingivalis and Treponema Denticola Exhibit Metabolic Symbioses. Plos Pathogens 10:e1003955. doi:10.1371/journal.ppat.1003955.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedKlein - ↑ 11.0 11.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedNaito - ↑ 12.0 12.1 Genbank nucleotide sequence database. accession: AP009380.1: "Porphyromonas gingivalis ATCC 33277 DNA, complete genome"
This page is written by Ron Ramsay for the MICR3004 course, Semester 2, 2016

