Salmonella enterica NEU2011: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
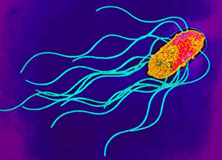
[[Image:S_enterica.JPEG|thumb|300px|right|Transmission Electron Micrograph of <i>Salmonella enterica</i>.Photogragh from [http://www.textbookofbacteriology.net/salmonella.html (1). Todar's Online Textbook of Bacteriology ]]] | [[Image:S_enterica.JPEG|thumb|300px|right|Transmission Electron Micrograph of <i>Salmonella enterica</i>.Photogragh from [http://www.textbookofbacteriology.net/salmonella.html (1). Todar's Online Textbook of Bacteriology ]]] | ||
==Classification== | ==Classification== | ||
Revision as of 21:41, 26 March 2011
Classification
Higher Order Taxa
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacteriales; Enterobacteriaceae; Salmonella; enterica.
Description and Significance
Salmonella is a gram-negative, rod-shaped, flagellated bacterium that is of interest due to its ability to cause infectious disease in humans and animals. Salmonella is facultative, being capable of both aerobic respiration for the production ATP as well as fermentation in the absence of oxygen. Dr. Daniel E. Salmon was the first person to identify Salmonella over 100 years ago, currently there are approximately 2,300 serotypes known. Salmonella infection is known as Salmonellosis; Salmonella enterica serovar thyphi is a subspecies of Salmonella that is responsible for typhoid fever in humans by invading the gastrointestinal tract. Much of the current understanding of cellular and molecular functions in a cell has been made possible by using Salmonella. For example, Salmonella enterica serovar Typhimurium, a subspecies of Salmonella enterica, was the first bacteria used to observe transduction by Zinder and Lederberg in 1952. Since then a vast number of discoveries have been made using Salmonella ranging from gene regulation mechanisms, to cell surface antigen interactions with immune cells Todar (2). (2).
Genome Structure
The genome of Salmonella enterica serovar Typhimurium LT2 is approximately 4,857 kilobases long and an additional 94 kilobase virulent plasmid specific to the LT2 strain. Lateral genetic transfer is common as 11% of the genes present in LT2 are missing from Salmonella enterica serovar typhi and 29% of genes are missing from Escherichia coli K12."
Cell Structure and Metabolism
Ecology
Pathology
Infectious diseases caused by Salmonella enterica depend largely on secreted proteins and adhesions from both fimbrial and non-fimbrial sources to create a biofilm and contact with the host cell (Hensel, 2009). Bacterial cells enter the cells that line the intestine, and the host cell’s membrane ruffles as a response to the initial attachment of the bacteria. Ruffling is related to a triggering reaction that is results in macropinocytosis. The vesicles are fairly large during macropinocytosis, providing an efficient route for non-selective endocytosis of solute macromolecules (Swanson and Watts 1995). The entrance of the bacterial cells briefly harms the microvilli on the cell surface. Following this disruption, white blood cells flood the mucosa which alters the homeostasis between absorption and secretion in the body (McCormick, 2007).
Bacterial cells harm the host cells by causing intracellular unrestricted calcium to rise and dislocate the cytoplasm. The infectious cells can then be transferred to the liver or spleen, where it multiplies (Giannella, 1996). S.enterica either return to the host’s intestinal tract, or can be defecated. In addition to food contamination, further infection often occurs from infested feces by polluted water, soil, or other unclean environments.
Salmonella enterica is the cause of many cases of abdominal complications like gastroenteritis. When the balance between absorption and secretion is altered, initial symptoms include abdominal cramps, diarrhea, fever, and nausea. Most cases caused by food contamination resolve themselves within one week’s time, but more serious cases can often need fluoroquinolone or cephalosporins, both are broad-spectrum antibiotics that can respectively kill the organism or stop the cell wall from forming (CDC).
Current Research
Cool Factor
References
1. Todar, Kenneth. 2008. Salmonella and Salmonellosis. Todar’s Online Textbook of Bacteriology
Mobley, Harry LT, George L Mendz,2and Stuart L Hazell. 2001. Helicobacter pylori Physiology and Genetics. Washington, DC: ASM Press
McClelland, Michael , et al. “Complete genome sequence of Salmonella enterica serovar Typhimurium LT2”. Nature. 2001. Volume 413. P. 852-856.
Edited by Abigail Bootes, Matthew Brennan, Alex Birch, and Danielle Laramee for BIOL2321 Microbiology, 2011, Northeastern University
