Simian Immunodeficiency Virus – Research subjects for vaccine and drug development and models for the origin of HIV and AIDS: Difference between revisions
| Line 25: | Line 25: | ||
Molecular efforts in dating the age of the SIV, or more precisely, the primate lentivirus lineage, based on the modeled mutation rate of HIV-1 and HIV-2 (the most well-studied primate lentiviruses) have bred great controversies. Dating methods using nucleotide molecular clocks that use the mutation rate of HIV-1 and HIV-2 as standard references suggest the most recent common ancestor of the primate lentivirus to be around a few hundreds to a few thousands of years before present<ref>Wertheim, Joel O., and Michael Worobey. 2009. “Dating the Age of the SIV Lineages That Gave Rise to HIV-1 and HIV-2.” PLoS Computational Biology 5(5):e1000377–e1000377.</ref><ref name = Sharp2000/>. This contradicts with the pathological and epidemiological evidences suggesting SIV might have evolved in a host-dependent manner to be non-pathogenic in some African monkeys <ref name = Sharp2010/><ref name = Santiago/><ref name = Peeters/><ref name = Beer/>. However, a study in 2010<ref name = Worobey2010/> combining the viruses’ molecular clock dating and phylogeny analysis with the host species’ phylogenic studies and biogeographical history suggests that the SIV lineage have existed for at least 32,000 years. Another research into the existence of SIV in the ring-tailed lemur of south-western Madagascar showed the existence of antigens against SIV in ring-tailed lemurs here. If there was no cross-contamination of SIV from mainland and SIV in primates evolved in a host-dependent way from a common ancestor (as seen in the case of green monkeys (<i>Chlorocebus</i> sp.)<ref name = Beer/>, this can push the possible date of the oldest common ancestor of the SIV lineage to 85 million years ago, the approximate date of the last primate common ancestor, also around the time of the total separation of Madagascar from the Indian continent<ref>Goodman, Morris, Danilo A. Tagle, David H. A. Fitch, Wendy Bailey, John Czelusniak, Ben F. Koop, Philip Benson, and Jerry L. Slightom. 1990. “Primate Evolution at the DNA Level and a Classification of Hominoids.” Journal of Molecular Evolution 30(3):260–66.</ref>. | Molecular efforts in dating the age of the SIV, or more precisely, the primate lentivirus lineage, based on the modeled mutation rate of HIV-1 and HIV-2 (the most well-studied primate lentiviruses) have bred great controversies. Dating methods using nucleotide molecular clocks that use the mutation rate of HIV-1 and HIV-2 as standard references suggest the most recent common ancestor of the primate lentivirus to be around a few hundreds to a few thousands of years before present<ref>Wertheim, Joel O., and Michael Worobey. 2009. “Dating the Age of the SIV Lineages That Gave Rise to HIV-1 and HIV-2.” PLoS Computational Biology 5(5):e1000377–e1000377.</ref><ref name = Sharp2000/>. This contradicts with the pathological and epidemiological evidences suggesting SIV might have evolved in a host-dependent manner to be non-pathogenic in some African monkeys <ref name = Sharp2010/><ref name = Santiago/><ref name = Peeters/><ref name = Beer/>. However, a study in 2010<ref name = Worobey2010/> combining the viruses’ molecular clock dating and phylogeny analysis with the host species’ phylogenic studies and biogeographical history suggests that the SIV lineage have existed for at least 32,000 years. Another research into the existence of SIV in the ring-tailed lemur of south-western Madagascar showed the existence of antigens against SIV in ring-tailed lemurs here. If there was no cross-contamination of SIV from mainland and SIV in primates evolved in a host-dependent way from a common ancestor (as seen in the case of green monkeys (<i>Chlorocebus</i> sp.)<ref name = Beer/>, this can push the possible date of the oldest common ancestor of the SIV lineage to 85 million years ago, the approximate date of the last primate common ancestor, also around the time of the total separation of Madagascar from the Indian continent<ref>Goodman, Morris, Danilo A. Tagle, David H. A. Fitch, Wendy Bailey, John Czelusniak, Ben F. Koop, Philip Benson, and Jerry L. Slightom. 1990. “Primate Evolution at the DNA Level and a Classification of Hominoids.” Journal of Molecular Evolution 30(3):260–66.</ref>. | ||
These directs and indirect evidence of SIV’s historic existence among the primates, especially those of the more ancient orders (monkeys and lemurs) who SIV have evolved natural co-existing relationships with, suggest a terrible bias in the HIV-based molecular clock assay, which points to a possible, drastic increase in HIV comparing to their more ancient SIV counterparts. On the other hand, it’s also HIV’s notoriously rapid mutation rate that is the main obstacles against development of vaccines against HIV and the primary cause of drug resistance in treatments. Thus, further research into the infections and mutating and replicating activities of older, naturally non-pathogenic SIV species can help us understand the genetic difference that enables the rapid mutation rate in their human-infecting counterparts, as well as develop vaccines against HIV. Moreover, considering the long existence of SIV (at least 32,000 years ago), it is likely that human has come into contact with SIV multiple times and cross-transmission of SIV to human are likely to have occurred many times throughout the past 32,000 years. Thus, there must be recent social and biological factors that the rapid spread of HIV only during the past 100 years or so | These directs and indirect evidence of SIV’s historic existence among the primates, especially those of the more ancient orders (monkeys and lemurs) who SIV have evolved natural co-existing relationships with, suggest a terrible bias in the HIV-based molecular clock assay, which points to a possible, drastic increase in HIV comparing to their more ancient SIV counterparts. On the other hand, it’s also HIV’s notoriously rapid mutation rate that is the main obstacles against development of vaccines against HIV and the primary cause of drug resistance in treatments. Thus, further research into the infections and mutating and replicating activities of older, naturally non-pathogenic SIV species can help us understand the genetic difference that enables the rapid mutation rate in their human-infecting counterparts, as well as develop vaccines against HIV. Moreover, considering the long existence of SIV (at least 32,000 years ago), it is likely that human has come into contact with SIV multiple times and cross-transmission of SIV to human are likely to have occurred many times throughout the past 32,000 years. Thus, there must be recent social and biological factors that the rapid spread of HIV only during the past 100 years or so<ref>Lemey, Philippe, Oliver G. Pybus, Bin Wang, Nitin K. Saksena, Marco Salemi, and Anne-Mieke Vandamme. 2003. “Tracing the Origin and History of the HIV-2 Epidemic.” Proceedings of the National Academy of Sciences of the United States of America 100(11):6588–92.</ref><ref>Worobey, Michael, Marlea Gemmel, Dirk E. Teuwen, Tamara Haselkorn, Kevin Kunstman, Michael Bunce, Jean-Jacques Muyembe, Jean-Marie M. Kabongo, Raphaël M. Kalengayi, Eric Van Marck, M. Thomas P. Gilbert, and Steven M. Wolinsky. 2008. “Direct Evidence of Extensive Diversity of HIV-1 in Kinshasa by 1960.” Nature 455(7213):661–64.</ref>. | ||
==Section 4== | ==Section 4== | ||
Revision as of 03:11, 25 April 2020
Overview

Simian immunodeficiency viruses (SIV) are a group within the Lentiviridae genus of retrovirus family, (Retroviridae) whose genome is ribonucleic acid (RNA) but reverse transcribe to deoxyribonucleic acid (DNA) intermediates before making viral proteins. This viral group is found naturally infecting more than 40 African monkeys (or Old World monkeys – Cercopithecidae family) and 2 species of great apes, namely chimpanzee (Pan troglodytes) and gorilla (Gorilla gorilla)[3]. SIV are the most closely related viruses to the human immunodeficiency viruses type 1 and 2 (HIV–1 and HIV–2), which cause acquire immunodeficiency syndrome (AIDS) in human[4]. Despite being called “immunodeficiency viruses”, SIV do not usually result in any AIDS-like syndromes in many of their natural African monkey host communities, such as sooty mangabeys (Cercocebus atys) and Colobus monkeys (Colobinae subfamily) even with high viral counts in infected individuals and high prevalence of infection (up to 59% of a community can be infected)[2][5][6][7]. This suggests that SIV have a long history of coevolution with these natural hosts. However, there has evidence suggesting recent host jumps of SIV from their long-adapted hosts to new host species such as chimpanzees in the wilds, macaques in captives (an example of Asian monkeys who have much less exposure to SIV than the African ones) and humans, where they developed into new strands through combinations and mutations and became pathogenic in the new host [8][9][10][11]. In macaques and chimpanzees, (SAIDS) similar to AIDS in human[8][12]. Thus, researches into SIV, in both their natural reservoir and laboratory model primates, not only reveals the origin of HIV and AIDS, but also help further understandings in HIV’s pathology and developments of HIV and AIDS prevention and treatments.
Taxonomy
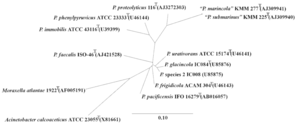
SIV are defined as lentiviruses (which itself belongs to the subfamily Orthoretrovirinae in the Retroviridae virus family) that specifically infect non-human primates (simians). The Lentiviridae genus includes a wide variety of primate infecting viruses and a few other viral species that infect felines, cows, horses, goats, and sheep[19][6][20]. Infection of lentiviruses are characterized by long dormant periods (infection may never progress to disease phase as in the case of some natural host monkeys), progressive disease development, permanent damage of tissues, organs, or systems, and, in many cases, lethal outcomes[19]. SIV are a polyphyletic group in the primate lentivirus that also include HIV-1 and HIV-2 (Figure 2)[4]. The SIV species in chimpanzee (SIVcpz) is closely related to the HIV-1 groups M and N[21], while HIV-1 group O is more closely related to the SIV species in gorilla (SIVgor)[22]. And the SIV species that is naturally non-pathogenic in sooty mangabeys (SIVsm) has been shown to be ancestral to HIV-2 and SIV species that cause lethal SAIDS in Asian macaque monkey species[23]. A full phylogeny of SIV along with different types of HIV and other non-primate lentiviruses are demonstrated in Figure 2 (species relatedness was determined by the molecular clock gene pol)[4].
Structure - SIV in non-human primates as an effective and accessible animal model for HIV research
SIV share characteristic traits in morphology and morphogenesis with other member of the lentivirus genus[24]. Genetically, the closest-related viral species to SIV are HIV-1 and HPV-2[21][22][23], the other two members of the primate lentivirus subgenus[4] and the most well-studied species among lentiviruses. Thus, according to a model of lentivirus structured based on HIV-1 and HIV-2, a SIV virion outside of a host cell, along with other lentiviruses’ virions, are either spherical or pleomorphic. SIV virion likely has an outer-membrane derived from host protein with viral spike proteins that binds specifically with host species of each SIV species. Inside the outer-membrane is a protein capsid containing a tightly coiled positive (+) single-stranded RNA genome and a few crucial viral proteins for controlling the host cell’s defense system, reverse transcription, and integration into host genome, such as reverse transcriptase, intergrase, and tRNA lysase. Once inside host cells, SIV and other lentiviruses do not exist as distinct virus particles, but reverse transcribe their RNA genomes into double-stranded DNA and integrate into the host genome with finesse to not cause any lethal mutations or trigger any oncogenes in host cells. Then, the viral double-stranded DNA would exploit the host’s transcribing and translating system to transcribe viral proteins and assemble and disperse progeny virions[25].
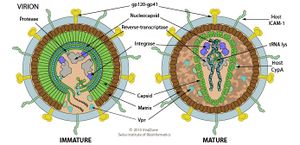
SIV in non-human primates as an effective and accessible animal model for HIV research
Due to the close resemblance and relatedness of SIV to HIV, the most similar anatomies of SIV’s hosts (non-human primates) and human, and the ability of some SIV speciess to induce simian AIDS in the common primate model organisms, the Asian macaques (Macaca spp.), with rapid disease progression and pathology similar to human AIDS, SIV effectively serves as models to study HIV and AIDS in animals[26][12][10]. However, the SIV species that infect Asian macaques are in the lineage SIVsmm/HIV-2 (Figure 2), which is relatively distant from the common global pandemic HIV-1 species, the macaques model still has certain differences in disease progression and drug response from the more common HIV-1[26] that researchers need to keep in mind when adopting this model organism for HIV researches. On the other hand, the closest animal model to HIV-1, and probably HIV-1-caused AIDS, has been shown to the SIVcpz found in the natural reservoir of central and eastern chimpanzee subspecies (Pan troglodytes troglodytes and Pan troglodytes schweinfurthii respectively), each of them have their own distinct type of SIVcpz infecting only that on subspecies[2]. Previously, all SIV found in natural reservoirs were not thought to be pathogenic, however, fecal sampling for SIV antibodies and behavioral studies in feral chimpanzees revealed that SIVcpz infection in wild chimpanzees can be pathologic progress to SAIDS with histopathology similar to what is seen in human’s AIDS[8]. Moreover, transmission of SIVcpz in the wild is also well-documented to be mostly sexual transmission and mother-to-infants transmission similar to what is observed in humans[8][27]. One limit to this SIV model is that chimpanzees (Pan troglodytes) and their counterparts, the bonobos (Pan paniscus), are rare, endangered species, thus limiting researchers’ ability to study them in laboratory settings. However, chimpanzee species and subspecies are the best-studied primate species in the wild with many groups studying chimpanzee and bonobos communities for multiple decades[28]. Thus, studying virology and pathology of SIVcpz in the wild and documenting behaviors and social interactions of infected chimpanzee communities in the wild can produce great insights into the virology, transmission, pathology, and social factors related to transmission of HIV-1.
Evolution of SIV – The origin of Human Immunodeficiency Virus (HIV) from SIV
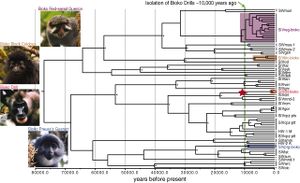
Molecular efforts in dating the age of the SIV, or more precisely, the primate lentivirus lineage, based on the modeled mutation rate of HIV-1 and HIV-2 (the most well-studied primate lentiviruses) have bred great controversies. Dating methods using nucleotide molecular clocks that use the mutation rate of HIV-1 and HIV-2 as standard references suggest the most recent common ancestor of the primate lentivirus to be around a few hundreds to a few thousands of years before present[30][9]. This contradicts with the pathological and epidemiological evidences suggesting SIV might have evolved in a host-dependent manner to be non-pathogenic in some African monkeys [2][5][6][7]. However, a study in 2010[29] combining the viruses’ molecular clock dating and phylogeny analysis with the host species’ phylogenic studies and biogeographical history suggests that the SIV lineage have existed for at least 32,000 years. Another research into the existence of SIV in the ring-tailed lemur of south-western Madagascar showed the existence of antigens against SIV in ring-tailed lemurs here. If there was no cross-contamination of SIV from mainland and SIV in primates evolved in a host-dependent way from a common ancestor (as seen in the case of green monkeys (Chlorocebus sp.)[7], this can push the possible date of the oldest common ancestor of the SIV lineage to 85 million years ago, the approximate date of the last primate common ancestor, also around the time of the total separation of Madagascar from the Indian continent[31].
These directs and indirect evidence of SIV’s historic existence among the primates, especially those of the more ancient orders (monkeys and lemurs) who SIV have evolved natural co-existing relationships with, suggest a terrible bias in the HIV-based molecular clock assay, which points to a possible, drastic increase in HIV comparing to their more ancient SIV counterparts. On the other hand, it’s also HIV’s notoriously rapid mutation rate that is the main obstacles against development of vaccines against HIV and the primary cause of drug resistance in treatments. Thus, further research into the infections and mutating and replicating activities of older, naturally non-pathogenic SIV species can help us understand the genetic difference that enables the rapid mutation rate in their human-infecting counterparts, as well as develop vaccines against HIV. Moreover, considering the long existence of SIV (at least 32,000 years ago), it is likely that human has come into contact with SIV multiple times and cross-transmission of SIV to human are likely to have occurred many times throughout the past 32,000 years. Thus, there must be recent social and biological factors that the rapid spread of HIV only during the past 100 years or so[32][33].
Section 4
Conclusion
References
- ↑ Murphey-Corb, Michael, Louis N. Martin, S. R. S. Rangan, Gary B. Baskin, Bobby J. Gormus, Robert H. Wolf, W. Abe Andes, Melanie West, and Ronald C. Montelaro. 1986. “Isolation of an HTLV-III-Related Retrovirus from Macaques with Simian AIDS and Its Possible Origin in Asymptomatic Mangabeys.” Nature 321(6068):435–37.
- ↑ 2.0 2.1 2.2 2.3 Sharp, Paul M., and Beatrice H. Hahn. 2010. “The Evolution of HIV-1 and the Origin of AIDS.”
- ↑ Sharp, Paul M., and Beatrice H. Hahn. 2011a. “Origins of HIV and the AIDS Pandemic.” Cold Spring Harbor Perspectives in Medicine 1(1):a006841.
- ↑ 4.0 4.1 4.2 4.3 4.4 Perrone, Rosalba, Enrico Lavezzo, Giorgio Palu, and Sara Richter. 2017. “Conserved Presence of G-Quadruplex Forming Sequences in the Long Terminal Repeat Promoter of Lentiviruses.” Scientific Reports 7.
- ↑ 5.0 5.1 Santiago, Mario L., Friederike Range, Brandon F. Keele, Yingying Li, Elizabeth Bailes, Frederic Bibollet-Ruche, Cecile Fruteau, Ronald Noë, Martine Peeters, John F. Y. Brookfield, George M. Shaw, Paul M. Sharp, and Beatrice H. Hahn. 2005. “Simian Immunodeficiency Virus Infection in Free-Ranging Sooty Mangabeys (Cercocebus Atys Atys) from the Taï Forest, Côte d’Ivoire: Implications for the Origin of Epidemic Human Immunodeficiency Virus Type 2.” Journal of Virology 79(19):12515–27.
- ↑ 6.0 6.1 6.2 Peeters, Martine, and Valerie Courgnaud. 2002. “Overview of Primate Lentiviruses and Their Evolution in Non-Human Primates in Africa.” HIV Sequence Compendium.
- ↑ 7.0 7.1 7.2 Beer, Brigitte E., Elizabeth Bailes, Robert Goeken, George Dapolito, Cheik Coulibaly, Stephen G. Norley, Reinhard Kurth, Jean-Pierre Gautier, Annie Gautier-Hion, and Dominique Vallet. 1999. “Simian Immunodeficiency Virus (SIV) from Sun-Tailed Monkeys (Cercopithecus Solatus): Evidence for Host-Dependent Evolution of SIV within the C. LhoestiSuperspecies.” Journal of Virology 73(9):7734–44.
- ↑ 8.0 8.1 8.2 8.3 Keele, Brandon F., James Holland Jones, Karen A. Terio, Jacob D. Estes, Rebecca S. Rudicell, Michael L. Wilson, Yingying Li, Gerald H. Learn, T. Mark Beasley, Joann Schumacher-Stankey, Emily Wroblewski, Anna Mosser, Jane Raphael, Shadrack Kamenya, Elizabeth V Lonsdorf, Dominic A. Travis, Titus Mlengeya, Michael J. Kinsel, James G. Else, Guido Silvestri, Jane Goodall, Paul M. Sharp, George M. Shaw, Anne E. Pusey, and Beatrice H. Hahn. 2009. “Increased Mortality and AIDS-like Immunopathology in Wild Chimpanzees Infected with SIVcpz.” Nature 460(7254):515–19.
- ↑ 9.0 9.1 Sharp, P. M., E. Bailes, F. Gao, B. E. Beer, V. M. Hirsch, and B. H. Hahn. 2000. “Origins and Evolution of AIDS Viruses: Estimating the Time-Scale.” Biochemical Society Transactions 28(2):275–82.
- ↑ 10.0 10.1 Gardner, Murray B., and Paul A. Luciw. 1989. “Animal Models of AIDS.” The FASEB Journal 3(14):2593–2606.
- ↑ Barré-Sinoussi, Françoise. 1996. “HIV as the Cause of AIDS.” The Lancet 348(9019):31–35.
- ↑ 12.0 12.1 Letvin, N. L., K. A. Eaton, W. R. Aldrich, P. K. Sehgal, B. J. Blake, S. F. Schlossman, N. W. King, and R. D. Hunt. 1983. “Acquired Immunodeficiency Syndrome in a Colony of Macaque Monkeys.” Proceedings of the National Academy of Sciences of the United States of America 80(9):2718–22.
- ↑ http://www.freepik.com/free-vector/business-team-outlines-pack_831669.htm#term=human
- ↑ http://www.flaticon.com/free-icon/monkey_47138
- ↑ http://www.freepik.com/free-vector/cat-silhouettes-set_718091.htm#term=cat
- ↑ http://www.flaticon.com/free-icon/horse-standing-black-shape_35907
- ↑ http://www.freepik.com/free-vector/cows-and-bull-silhouettes_788343.htm
- ↑ http://www.freepik.com/free-vector/pack-of-farm-animal-silhouettes_1058750.htm#term=sheep&page=1&position=29
- ↑ 19.0 19.1 Shuljak, B. F. 2006. “Lentiviruses in Ungulates. I. General Features, History and Prevalence.” Bulgarian Journal of Veterinary Medicine 9(3):175–81.
- ↑ Pedersen, N. C., J. K. Yamamoto, T. Ishida, and H. Hansen. 1989. “Feline Immunodeficiency Virus Infection.” Veterinary Immunology and Immunopathology 21(1):111–29.
- ↑ 21.0 21.1 Keele, Brandon F., Fran Van Heuverswyn, Yingying Li, Elizabeth Bailes, Jun Takehisa, Mario L. Santiago, Frederic Bibollet-Ruche, Yalu Chen, Louise V Wain, Florian Liegeois, Severin Loul, Eitel Mpoudi Ngole, Yanga Bienvenue, Eric Delaporte, John F. Y. Brookfield, Paul M. Sharp, George M. Shaw, Martine Peeters, and Beatrice H. Hahn. 2006. “Chimpanzee Reservoirs of Pandemic and Nonpandemic HIV-1.” Science (New York, N.Y.) 313(5786):523–26.
- ↑ 22.0 22.1 Pedersen, N. C., J. K. Yamamoto, T. Ishida, and H. Hansen. 1989. “Feline Immunodeficiency Virus Infection.” Veterinary Immunology and Immunopathology 21(1):111–29.
- ↑ 23.0 23.1 Apetrei, Cristian, Amitinder Kaur, Nicholas W. Lerche, Michael Metzger, Ivona Pandrea, Johnny Hardcastle, Shelley Falkenstein, Rudolf Bohm, Jeffrey Koehler, Vicki Traina-Dorge, Tessa Williams, Silvija Staprans, Gail Plauche, Ronald S. Veazey, Harold McClure, Andrew A. Lackner, Bobby Gormus, David L. Robertson, and Preston A. Marx. 2005. “Molecular Epidemiology of Simian Immunodeficiency Virus SIVsm in U.S. Primate Centers Unravels the Origin of SIVmac and SIVstm.” Journal of Virology 79(14):8991.
- ↑ Desrosiers, Ronald C. 1990. “The Simian Immunodeficiency Viruses.” Annual Review of Immunology 8(1):557–78.
- ↑ https://viralzone.expasy.org/264?outline=all_by_species
- ↑ 26.0 26.1 Policicchio, Benjamin B., Ivona Pandrea, and Cristian Apetrei. 2016. “Animal Models for HIV Cure Research.” Frontiers in Immunology 7:12.
- ↑ Rudicell, Rebecca S., James Holland Jones, Emily E. Wroblewski, Gerald H. Learn, Yingying Li, Joel D. Robertson, Elizabeth Greengrass, Falk Grossmann, Shadrack Kamenya, and Lilian Pintea. 2010. “Impact of Simian Immunodeficiency Virus Infection on Chimpanzee Population Dynamics.” PLoS Pathogens 6(9).
- ↑ Pusey, Anne E., Lilian Pintea, Michael L. Wilson, Shadrack Kamenya, and Jane Goodall. 2007. “The Contribution of Long‐term Research at Gombe National Park to Chimpanzee Conservation.” Conservation Biology 21(3):623–34.
- ↑ 29.0 29.1 Worobey, Michael, Paul Telfer, Sandrine Souquière, Meredith Hunter, Clint A. Coleman, Michael J. Metzger, Patricia Reed, Maria Makuwa, Gail Hearn, Shaya Honarvar, Pierre Roques, Cristian Apetrei, Mirdad Kazanji, and Preston A. Marx. 2010. “Island Biogeography Reveals the Deep History of SIV.” Science 329(5998):1487.
- ↑ Wertheim, Joel O., and Michael Worobey. 2009. “Dating the Age of the SIV Lineages That Gave Rise to HIV-1 and HIV-2.” PLoS Computational Biology 5(5):e1000377–e1000377.
- ↑ Goodman, Morris, Danilo A. Tagle, David H. A. Fitch, Wendy Bailey, John Czelusniak, Ben F. Koop, Philip Benson, and Jerry L. Slightom. 1990. “Primate Evolution at the DNA Level and a Classification of Hominoids.” Journal of Molecular Evolution 30(3):260–66.
- ↑ Lemey, Philippe, Oliver G. Pybus, Bin Wang, Nitin K. Saksena, Marco Salemi, and Anne-Mieke Vandamme. 2003. “Tracing the Origin and History of the HIV-2 Epidemic.” Proceedings of the National Academy of Sciences of the United States of America 100(11):6588–92.
- ↑ Worobey, Michael, Marlea Gemmel, Dirk E. Teuwen, Tamara Haselkorn, Kevin Kunstman, Michael Bunce, Jean-Jacques Muyembe, Jean-Marie M. Kabongo, Raphaël M. Kalengayi, Eric Van Marck, M. Thomas P. Gilbert, and Steven M. Wolinsky. 2008. “Direct Evidence of Extensive Diversity of HIV-1 in Kinshasa by 1960.” Nature 455(7213):661–64.
Authored by [Minh Pham] for BIOL 238 Microbiology, taught by Joan Slonczewski, 2020, Kenyon College.
