Sporomusa silvacetica
Classification
Kingdom: Bacteria
Phylum: Firmicutes
Class: Negativicutes
Order: Selenomonadales
Family: Veillonellaceae
Genus: Sporomusa
Species: Sporomusa silvacetica
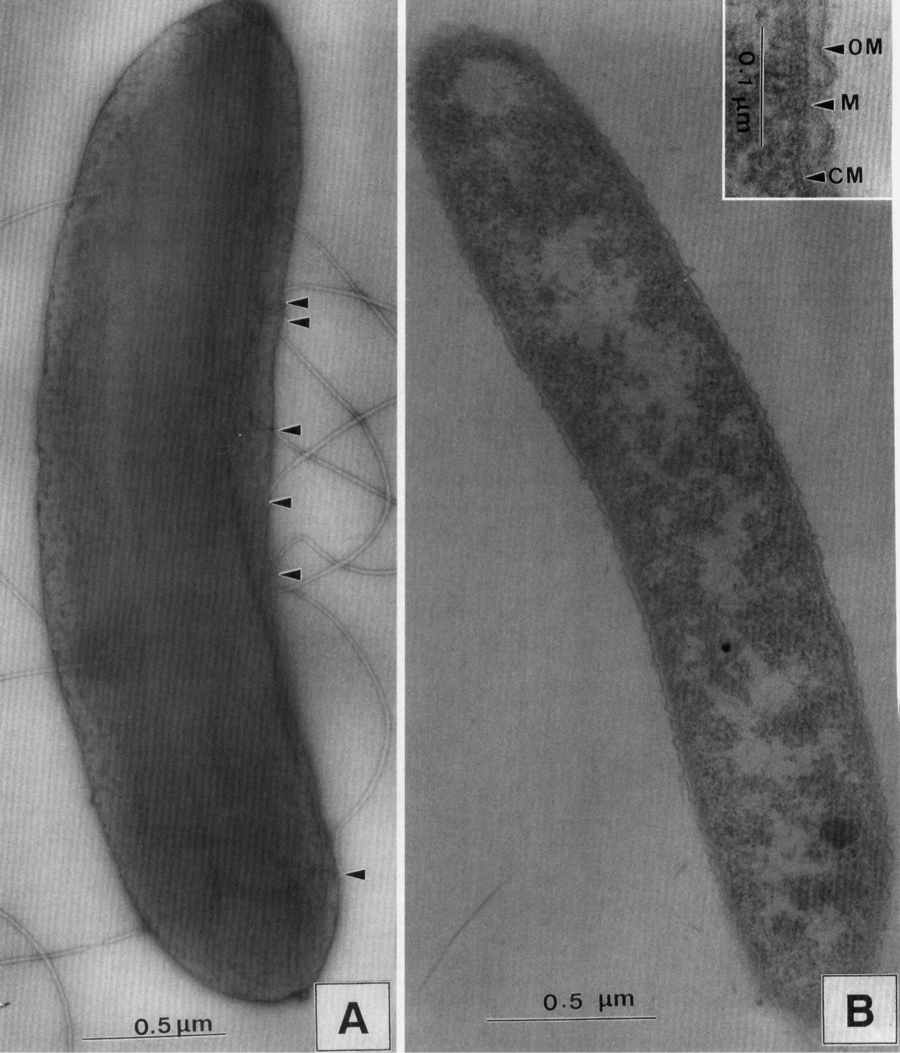 Transmission electron micrographs of a negatively-stained Sporomusa silvacetica[2].
Transmission electron micrographs of a negatively-stained Sporomusa silvacetica[2].
Species
|
NCBI: Taxonomy |
Sporomusa silvacetica
The genus name Sporomusa means “spore-bearing banana,” which describes the slightly curvy rod shaped cells of bacteria belonging to this particular genus.
Description and Significance
Sporomusa silvacetica was first discovered in the late 1990s in Geisberger Forest in east-central Germany. Kuhner and his team isolated the bacteria from a sample of drained forest soil. Like all bacteria belonging to the genus Sporomusa, Sporomusa silvacetica has curvy rod shaped cells and produces spherical shaped spores. The approximate cell size was measured as 3.5 by 0.7 µm. The bacterial cells are motile by means of a flagellum for propulsion, which is located on the concave side of the cell. Sporomusa silvacetica has a multilayered cell wall, however the individual layers that make up the cell wall are not very thick. A Type c cytochrome was detected in the membrane, which was significant in determining the species. Consequently, Gram staining causes the bacteria to stain weakly Gram-positive[2].
Sporomusa silvacetica has also been identified as an anaerobe and homoacetogen. Consequently, the organism catalyzes acetate for energy to be used in its metabolism for growth[5]. Growing conditions of 25-30°C and pH 6.8 were defined as the optimal conditions for Sporomusa silvacetica. Doubling time of the bacteria was 14 hours when grown under these conditions and fructose was used as the substrate. Sporomusa silvacetica colonies grown on fructose are shiny beige yellow and approximately 2-3 mm in diameter. The bacteria are also capable at growing at temperatures as low as 5-10°C.
Genome Structure
This genome has not been sequenced, therefore the protein coding and other operations of the genome are speculative.
16s Ribosomal DNA & Comparisons
Approximately 95% of the 16s rRNA gene (rDNA) sequence of strain DG-1T was determined by directly sequencing PCR-amplified 16s rDNA[2]. Phylogenetic analysis using 16S ribosomal DNA sequencing indicates that Sporomusa silvacetica and other members of the Sporomusa clade are closely related to the gram positive members of the class Clostridia[2]. The cell shape, gram-positive reaction, formation of spores, and lack of sulfate reduction characteristics of Sporomusa silvacetica relate to the genus Clostridium[2]. However, supporting the 16s rDNA sequencing, Sporomusa silvacetica is more closely related to the gram-negative, spore-forming characeristics of the genus Sporomusa in the Sporomusa subranch of the Clostridium subphylum of the gram-positive bacteria[2]. This is interesting because it is still argued Sporomusa silvacetica belongs in the class Clostridia and not Negativiates. The similarities of the two genii and the characteristics of Sporomusa silvacetica make it difficult to identify the correct one, therefore a sequencing of the genome would be extremely helpful.
43 mol% of the Sporomusa silvacetica DNA consists of guanine and cytosine bases[2]. The guanine and cytosine count can be related to DNA stability, where the higher the count, the more stable the DNA. A 43 mol% guanine and cytosine count is significantly lower than other species, yet this trait belongs to a core group called the low-G+C group. Sporomusa silvacetica was placed in the phylum Firmicutes because this phylum belongs to this low-G+C group. Compared to the 47 mol% and 49 mol% of Sporomusa paucivorans and Sporomusa termitida, the low-G+C count of all three support the similarity of these species[2].
A type b cytochrome was detected in the membrane[2]. This complex is a part of the respiratory chain complex III and is involved in electron transport to create a proton motive force to generate ATP for necessary biological functions. This was helpful in determining the species of Sporomusa silvacetica because this type of cytochrome is a trait characteristic of other Sporomusa species, including both Sporomusa paucivorans and Sporomusa termitida[0]. Of the Sporomusa clade, Sporomusa paucivorans and Sporomusa termitida had similarity values of 97.2% and 95.9% when compared to Sporomusa silvacetica[2], most likely due to the similar type b cytochrome, the low-G+C count, and other cell structure similarities.
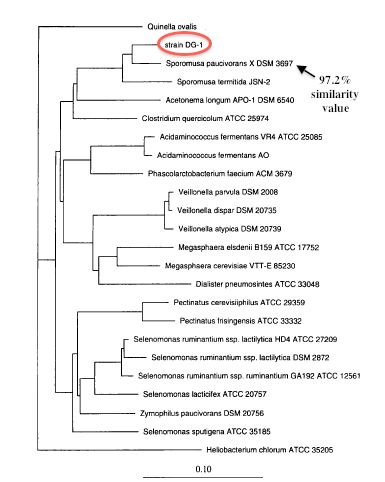 A dendrogram based on the 16s rDNA analysis shows the hierarchical clustering of Sporomusa[2].
A dendrogram based on the 16s rDNA analysis shows the hierarchical clustering of Sporomusa[2].
Metabolism
Sporomusa silvacetica is an anaerobic bacterium that uses acetogenesis for metabolism. Sporomusa silvacetica can utilize many different substrates in order to grow including ferulate, vanillate, fructose, betaine, fumarate, 2,3-butanediol, pyruvate, lactate, glycerol, ethanol, methanol, formate, and H2-CO2 [2]. Acetate is the primary reduced end product in most of the reactions using these substrates. This bacterium can use electron acceptors other than CO2 in order to conserve energy. Sporomusa silvacetica is the first discovered case in the Sporomusa genus to use fumarate and aromatic acrylate groups as terminal electron acceptors. It also can dismutate fumarate into succinate and acetate [2]. The acetyl-coenzyme A pathway is the primary pathway used when H2-CO2 is present [3]. Small traces of methane have also been produced when Sporomusa silvacetica utilizes H2-CO2. Neither nitrate nor sulfate was found to be the preferred terminal electron acceptor in any of these cases [2].
Oxygen
Oxygen can damage enzymes used in the acetyl-coenzyme A pathway in Sporomusa silvacetica. However, acetate production in Sporomusa silvacetica actually increases due to oxidative stress [1]. The amount of acetate production required for cell growth increases, and allows Sporomusa silvacetica to grow in environments that are not completely anaerobic. This is due to aerotolerance in the cell. Cell extracts of Sporomusa silvacetica contain various levels of perosidase to consume H202, and NADH oxidase to consume O2. The cell then rids itself of these toxic products [1].
Wood-Ljungdahl Pathway
When Sporomusa silvacetica grow in the presence of H2-CO2, the Wood-Ljungdahl Pathway is utilized within the cell [4]. This pathway is also called the acetyl-coenzyme A pathway. CO2 is converted into either cellular carbon, or acetyl phosphate and ATP. Carbon first enters the pathway at the CO2 reduction step and uses H2 as the electron donor. It is then reduced to CO. Acetyl–CoA is then produced by condensing a methyl group with CO using coenzyme A. Acetyl-CoA is then either used to make cellular carbon, or is converted into acetyl phosphate in order to make ATP and acetate for the cell [4].
Ecology
Sporomusa silvacetica is an organism that was isolated from well-drained soil in a beech forest east-central Germany. It uses H2-CO2 from the environment around it, and also utilizes its habitat for various sources of Carbon. It can adapt to environments that are strictly not anaerobic. It also utilizes H2 from various samples of tundra soils, forests, and prairies [1].
References
Author
Page authored by Bevneet Grewal, Jenna Mitchell, & Raajan Naik, students of Prof. Jay Lennon at Michigan State University.
