Streptococcus pneumoniae: Difference between revisions
| Line 14: | Line 14: | ||
''Streptococcus pneumoniae'' is a Gram-positive bacteria most widely known for its pathology. It is known to cause pneumonia, bacteremia, otitis media, and meningitis in humans. | ''Streptococcus pneumoniae'' is a Gram-positive bacteria most widely known for its pathology. It is known to cause pneumonia, bacteremia, otitis media, and meningitis in humans. | ||
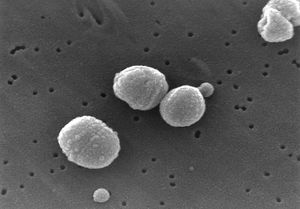
[[Image:Streptococcus_pneumoniae.jpg|right|thumbnail| | [[Image:Streptococcus_pneumoniae.jpg|right|thumbnail|Electron microscope photomicrograph of ''Streptococcus pneumoniae''. Source: | ||
CDC Public Health Image Library]] | |||
==Genome structure== | ==Genome structure== | ||
Revision as of 04:21, 6 June 2007
A Microbial Biorealm page on the genus Streptococcus pneumoniae
Classification
Higher order taxa
Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae
Species
Streptococcus pneumoniae
Description and significance
Streptococcus pneumoniae is a Gram-positive bacteria most widely known for its pathology. It is known to cause pneumonia, bacteremia, otitis media, and meningitis in humans.
Genome structure
The genome of "S. pneumoniae" consists of 2.16 million base pairs and contains 2236 predicted coding regions, 64% of which have been assigned a biological role. About 5% of the genome is made up of insertion sequences that may contribute to genome rearrangements through transferral of DNA. Several surface proteins have been identified that may serve as vaccine candidates. Several strains of "Streptococcus pneumoniae" have been identified, possibly accounting for differences in virulence and antigenicity.
Cell structure and metabolism
"Streptococcus pneumonia" gets a significant amount of its carbon and nitrogen through extracellular enzyme systems that allow the metabolism of polysaccharides and hexosamines, as well as cause damage to host tissue and enable colonization.
Ecology
Pathology
Streptococcus pneumoniae
Pneumonia is most prevalent in poorer countries, increasing the need for an economical and effective vaccine against Streptococcus pneumoniae. A potential candidate is the bacterium Lactococcus lactis, which produces the S. Pneumoniae surface protein A (PspA). Exposure to this strain may stimulate antibody production effective against S. Pneumoniae without running the risk of contracting the disease. (3)
Application to Biotechnology
Current Research
References
1. Hervé, T., et al. "Complete Genome Sequence of a Virulent Isolate of Streptococcus pneumoniae". Science. July 20, 2001. Volume 293. p. 498-506.
http://www.sciencemag.org/cgi/content/abstract/293/5529/498
2. Winstead, E. "Sugar Transporters and Foreign DNA, The sequenced Streptococcus pneumoniae genome". Genome News Network. July 23, 2001.
http://www.genomenewsnetwork.org/articles/07_01/Streptococcus_p.shtml
3. Hanniffy, S., Carter, A., Hitchin, E., and Wells, J. "Mucosal Delivery of a Pneumococcal Vaccine Using Lactococcus lactis Affords Protection against Respiratory Infection". The Journal of Infectious Diseases. 2007. Volume 195. p. 185-193.