Symbiotic relationship between Aliivibrio fischeri and Euprymna scolopes
By: Ben Hayden
Classification
Kingdom: Bacteria
Phylum: Proteobacteria
Class: Gamma Proteobacteria
Order: Vibrionales
Family: Vibrionaceae
Genus: Aliivibrio
Introduction
Aliivibrio spp. are gram-negative rod-shaped motile bacteria found in marine environments. Aliivibrio species are capable of bioluminescence and are known to form symbiotic relationships with marine organisms that predominantly exist in temperate waters. The origins of various luciferase systems evolved independently due to their ability to remove oxygen that was initially toxic to early life. As the abundance of oxygen grew the need for its removal became pointless, which led to the evolution of various uses of illumination ability. The abundance and distribution of Aliivibrio fischeri is also affected by its symbiotic relationship with Euprymna scolopes (11). In areas where the host is not present, symbiont populations average 30 times more than areas where the host exists.
Divergence with Vibrio Species
Previously recognized as Vibrio fischeri, this particular species group is now classified under the genus Aliivibrio to resolve prior ambiguity of the taxonomic and phylogenetic position of the clade (1). One notable conflict concerning the classification of Vibrio was to clarify differences between species that are pathogenic and species that are not (1). Though V. fischeri is not known to be pathogenic, strain E114 contains homologs of toxin encoding genes as it was initially a congeneric species with Vibrio. Among these are two CTX phage-encoding genes, zot (zona occludins toxin) and ace (accessory cholera enterotoxin). Ace has only been found in the gene sequences of V. fischeri and V. cholerae (24). An interesting difference between both species is the functional difference of another toxin termed RTX. In V. cholerae, the RTX protein impacts regulators of host’s actin polymerization which loosens the integrity of their tightly bound cytoskeleton. Though this still needs to be investigated, RTX of V. fischeri initiates a controlled change that deploys actin in the epithelial cells surrounding the light organ.
Multilocus phylogenetic analysis revealed a clear separation between Vibrio fischeri and other Vibrionaceae species. V. fischeri falls into a distinct clade that is related to Vibrio and Photobacterium but it is also dissimilar, even more dissimilar to Vibrionaceae species (1).
The Aliivibrio genus comprises four species: A. fischeri, A. salmonicida, A. wodanis, and A. logei. With regard to strains isolated from bioluminescent symbiosis, phylogenetic analysis reveals that these strains are discrete from Vibrio and other photobacterium (24). The four strains of Aliivibrio fischeri are: MJ1, ATCC7744, Y1, and ES114.
Cell Structure and Metabolism
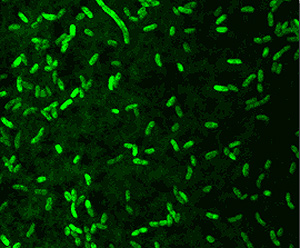
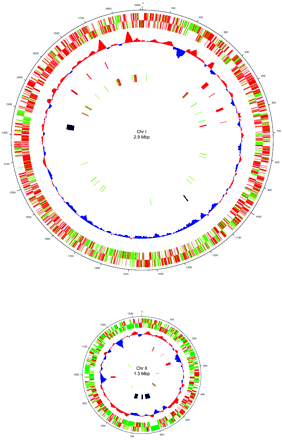
Aliivibrio fischeri is oxidase-positive facultative anaerobe and is capable of surviving on glucose as its only carbon source. The gram-negative rod-shaped motile bacteria contain a thick peptidoglycan layer is a necessary component in the recognition of a symbiotic host such as the Hawaiian bobtail squid, Euprymna scolopes. The genome is comprised of two circular chromosomes and a circular plasmid, pES100. It is comprised of There are twelve 16S ribosomal RNA operons, 11 of which reside on the larger chromosome (Chr I). Most of these operons are grouped in 3 loci. The remaining rrn operon exists on the smaller chromosome (Chr II) (24). These motile bacteria are driven by 1-3 unsheathed flagella and contain yellow cell-associated pigment. The G+C content is between 38.3 mol%. The size of the genomes contains 4,284,050 bps with an ORF number of 3,802. Within the 27 species of the Vibrionaceae family the A. fischeri genome has the lowest G-C content but is more closely related to the pathogenic Vibrio spp. with a higher G-C makeup (24). A. fischeri contain 10 separate pilus gene clustors and grow on media containing 1% NaCl, but not on 10% NaCl (1).
A. fischeri is a chemoorganotroph and its metabolism is dependant on its current state in the water column, whether it is free-living or symbiotic. Specifically in symbiosis, the bacterium is capable of metabolizing glucose and cyclic adenosine monophosphate (cAMP) (23). Within the host tissues, the metabolic mechanisms of A. fischeri are still uncertain and need to be monitored in situ. A symbiotic strain of A. fischeri is capable of growing in a medium of cAMP as its soul source for carbon and energy using active cAMP phosphodiesterase. In comparison to glucose-derived cultures, this strain had a slower growth rate, but a higher luminescence ability. When grown on glucose as a growth media A. fischeri produces a high quanity of pyruvate which can be utilized by the host cells to prevent acidification of the light organ (23).
Symbiotic Relationship
Aliivibrio species are capable of bioluminescence and through their symbiotic relationships with marine organisms they are capable of passing on this illumination ability to larger organisms. The ability for these animals to illuminate provides them with advantages such as: prey attraction, interspecific and intraspecific communication, predator deterrence and mate attraction.
Euprymna Scolopes
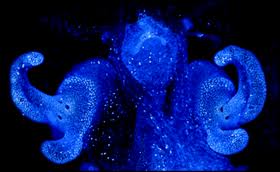
The model host for A. fischeri is the Hawaiian bobtail squid, Euprymna scolopes, which is capable of exploiting the bioluminescent properties of the marine bacteria. E. scolopes is a nocturnal organism that is native to the Pacific Ocean that inhabits shallow coastal waters in the photic zone and buries itself in the sand during the day. Transmission of this bacterium is done horizontally within a population; therefore A. fischeri cannot be inherited. Juvenile E. scolopes are aposymbiotic and needs to harvest the bacterium that it later stores in its light-emitting organ within the center of its mantel. As A. fischeri produce light, the organism is capable of controlling its glow, which it uses as a defense mechanism to avoid predators and to obtain prey (4). This method of camouflage is termed “counter-illumination” as it is capable of matching the illuminated sky when viewed from its ventral side. E. scolopes possess light receptors on their dorsal side that are capable of recognizing the light patterns emitted by the night sky. In order to replicate the patterns expressed by the stars, these organisms have appendages that act as shutters and allow the squid to monitor its bioluminescent display (13). The organ is continuously exposed to ambient saltwater and it contains specified tissues that provide optimal conditions for bacterial growth.
Mucus Secretion
E. scolopes hatchlings discharge a mucus into their immediate environment which is what attracts A. fischeri. Studies have dealt with the difference in bacterial presence within the host’s light organ crypts, which are extracellular epithelium-lined spaces (13). Hatchlings are constantly exposed to different mixtures of related gram-negative bacterial cells found in the natural habitat of E. scolopes including: A. fischeri, Vibrio parahaemolyticus or Photobacterium leiognathi. A replication experiment revealed that A. fischeri is the dominant species that accumulates in the crypts which suggest their binding affinity to the mucus is stronger than those of the latter two species (13). Dominance can also be attributed to the ability of A. fischeri cells to form tight populations.
Confocal microscopy analysis shows that light organ crypt initially acquire nonsymbiotic bacteria smaller than 2 micrometers in diameter during hour 1 after hatching (12). After 2 hours, these cells are no longer present and not able to enter the crypt. When the host is in contact with gram-negative bacteria the light organ’s superficial ciliated epithelium releases the mucus for 48 hours during the colonization of the symbiont until A. fischeri accumulation terminates the secretion (12). This discontinuation of the mucus indicates strong host-symbiont specificity between E. scolopes and A. fischeri.
Bacterial Colonization
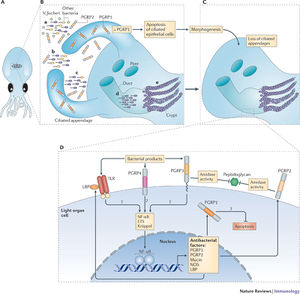
Seawater flows to the external pores of the light organ through a current driven by beating cilia. These pores lead to the three pairs of crypts inside the light organ, and once inside, A. fischeri populations reach 106 cells within 12 hours. As the animal develops, it is able to foster a population of 109 cells (20). There are two characteristics of this relationship that make it possible. First, the A. fischeri population stays monospecific, in that it avoids any potential contamination by different bacteria outside of the host. The second feature is the host-specificity of A. fischeri as it is not known to invade the tissues of other hosts.
The bacterial symbiont is linked to three types of host cells that either line the crypts or move about freely within the host. The two types of cells lining the crypt space are highly polarized epithelium cells and specialized goblet cells (20). Moving freely within this space are amoeboid granulocytes. The heterogeneous functions of these cells are key in the symbiont colonization. Once A. fischeri continue through the ducts, they bind to the epithelium cells lining the crypts and grow in biofilms until the space is filled (20). A daily circadian rhythm of E. scolopes is to expel 90-95% of the symbionts through its pores, retaining only 5-10% to establish a new population (13). Luminescence occurs once the symbionts reach a certain population size. The filtering of A. fischeri populations within these organisms then makes sense because E. scolopes live in the photic zone their luminescence is only necessary during the night.
The pores that lead to the crypts are open at all times, nonsymbiotic species of bacteria are able to access these areas of the host. The host uses phagocytes from the crypts as a defense mechanism to eliminate these bacteria. The function of these phagocytic cells are not well understood, however they are potentially key regulators of symbiont populations within the light organ (20).
Though the colonization of a symbiont is beneficial for the host, there are still physiological barriers that the bacteria need to overcome and in specifically in this relationship the bacterial cells must infiltrate the viscous barrier in order to access the crypts (8).
Developmental Changes
There are physiological changes that are associated with both organisms once the symbiotic relationship is established. In the first few hours of entering the host, A. fischeri lose their flagella. A quorum-sensing molecule known as VAI-1 accumulates in the crypts and increases the illumination ability of each cell 1000 fold (20).
E. scolopes also undergoes a morphological change as it loses both pairs of its ciliated appendages on either side of the light organ. These protrusions are known to assist with the colonization process and once the symbionts are retained and begin interacting with the crypts the light organ morphology changes. Once the population is established within the crypts this triggers programmed cell death which will eventually lead to the loss of their attachments (20). Within a couple days, the volume of epithelial cells quadruples.
Bioluminescence of A. fischeri
Bioluminescence has evolved separately in many different forms and therefore its regulation differs among the organisms that express it. In its most basic element, the emission of light during bioluminescence is the final product of a multistep exergonic reaction (28).
The light production of bioluminescence in A. fischeri is catalyzed by luciferase. This oxidation reaction occurs when an aliphatic aldehyde substrate, molecular oxygen, and a reduced flavin mononucleotide are present (20).
Tetradecanal + FMNH2 + O2 → Tetradecanoic acid + FMN + H2O + Light (490nm)
LuxAB genes encode for luciferase, luxCDE genes encode proteins for the biosynthesis of the aldehyde substrate and luxG is found downstream. Two regulatory lux genes reside on divergent operons adjacent to eachother (Ruby, 1996). The first of these regulatory genes produces LuxI which then synthesizes N-(3-oxohexanoyl) homoserine lactone which is termed the A. fischeri autoinducer (VAI-1) (20). The product of the latter regulatory gene, LuxR then binds to the autoinducer and activates the transcription of the lux operon. It is the lux operon that induces light producution 1000-1. VAI-2 is a subdued autoinducer of luminescence. Both autoinducers are produced in low concentrations and diffuse through the cell envelope. The optimal conditions for the induction of the lux operon are an enclosed environment with VAI-1 and a large population of A. fischeri. At the right concentrations of each they reach a state of equilibrium which generates the LuxR/VAI-1 complex, eventually inducing the lux operon. This process is known as quorum sensing in that bacteria have an awareness of conspecific cell density (3).
Modulation
A. fischeri have the ability to emit light constantly, which allows for host adaptations to regulate a bioluminescent display. A facilitated description of one process is similar to the light receptors in the eye. The crypts tissues surrounding the epithelia that help obtain the bacteria are very comparable to the tissue surrounding the retinas of the eye. As these tissues are able to soften the intensity of incoming light, the light organ of E. scolopes is able to moderate the light it that it emits (5). Lining the dorsal side of E. scolopes ink sac is a layered reflective tissue, which assures light exposure is strictly ventral. The ventral sac itself oscillates to permit specified light levels and because the ventral surface is covered with a muscle-driven lens it is able to diffuse bacterial luminescence (25). The second way in which the intensity of the symbiont’s light emission is controlled is by the host’s regulation of oxygen levels to the bacteria in its crypts (21). The luminescent properties of A. fischeri are driven by the oxidation of substrates. In limiting the amount of oxygen exposed to the bacteria, the host limits its luminescent activity. This modulation method of withholding oxygen from symbionts in the light organ has been well studied in anomaplopid and leiognathid fishes (21).
Cultured in Lab
When A. fischeri is cultured outside of the light organ, their maximum luminescent capability is reduced 1000-fold (4). The primary reason for this hindered light intensity is due to A. fischeri’s underproduction of luciferase autoinducer. This molecule is key in the positive transcriptional regulation of the bacteria’s lux operon, however optimal light intensity is also facilitated by the conditions provided by the host’s light organ (3).
This particular host-symbiont relationship is unique in that it is not a nutrition-based relationship. Because the neither provide essential nutrients for the other, they are able to be tested on in isolation. Under lab conditions, the host appears to be unaffected when A. fischeri is not present.
Other Aliivibrio Species
Based on the phylogenetic analysis of 16S rRNA gene sequences of the four strains of bacteria extracted from the Manila clam, Ruditapes philippinarum, most closely resembled genomes of Aliivibrio species (2). Gene sequence similarities ranged between 98.1 and 96.0% which brought about the proposed Aliivibrio finisterrensis sp. nov..
Aliivibrio salmonicida is a fish pathogen that is the causative agent of vibriosis in marine hatcheries. Symptoms of carrying the bacterium lead to hemolysis and tissue degradation in salmonids and Atlantic Cod (7).
Future Studies
Future work could look to understand how A. fischeri outcompete other bacteria in the mucus aggregations. When A. fischeri is not present, the mucus still obtains nonsymbiotic bacteria that then form fixed aggregations of the light organ. Therefore, the mucus is not completely selective to A. fischeri. Current data provides no evidence of higher symbiont growth or the death of nonsymbiont bacteria within the matrix (13).
Is there something particular about A. fischeri individuals that bind to the epithelial cells? As E. scolopes individuals are known to exude 90-95% of the A. fischeri symbionts out of their pores at dawn, the remaining 5-10% need to regenerate an entire population. Do the bacteria associated with the microvillus epithelium better off in order to assure a healthy population? Are the associations between host cells and symbionts random or are they selected for based on distinct characteristics?
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2014, Kenyon College.
References
3. Bonnie. 2009. How “bacteria talk”. TED2009.
6. Bretl, Todd. Underwater Photography. http://toddbretl.com/
15. McCapra, F. Chemical Mechanisms in Bioluminescence. Accounts of Chemical Research 9: 201-211.
16. McFall-Ngai, M.J. 2008. Hawaiian bobtail squid. Curr Biol. 18: 1043-1044.