The Threat of Airborne Transmission of Influenza A: H5N1: Difference between revisions
| Line 51: | Line 51: | ||
The shedding time of the influenza virus, or the duration of time in which an infected person might be infectious to others, begins about 24 hours before symptoms become overt (Hall et al., 2007). | The shedding time of the influenza virus, or the duration of time in which an infected person might be infectious to others, begins about 24 hours before symptoms become overt (Hall et al., 2007). | ||
The virus is released between 5 to 7 days, however cases may vary. Peak of infection is most often observed between day 2 and 3 (Hall et al., 2007). Shedding time of the virus is also correlated with temperature of the host environment as hosts with fevers usually have higher transmission rates (Weber et al., 2008). Children are also much more infectious than adults, as shedding time can last up to two weeks after initial infection (Weber et al., 2008). These are all important elements when taking into consideration how influenza might affect specific as well as general populations at a given time; mathematical models have also been generated to predict effects (Weber et al., 2008). | The virus is released between 5 to 7 days, however cases may vary. Peak of infection is most often observed between day 2 and 3 (Hall et al., 2007). Shedding time of the virus is also correlated with temperature of the host environment as hosts with fevers usually have higher transmission rates (Weber et al., 2008). Children are also much more infectious than adults, as shedding time can last up to two weeks after initial infection (Weber et al., 2008). These are all important elements when taking into consideration how influenza might affect specific as well as general populations at a given time; mathematical models have also been generated to predict effects (Weber et al., 2008). | ||
==Current Research== | ==Current Research== | ||
Revision as of 19:37, 23 April 2013
Introduction
The evolution of the influenza virus has long been a threat to our society. From the first major pandemic in 1918, involving the H1N1 "Spanish flu," which killed an estimated 50 million people and infected about 500 million people worldwide to the Swine flu in 2009 which affected one in six Americans, caused a reported 43-89 million cases and 8870-18300 deaths worldwide, viral reassortment and mutation have raised a serious global awareness in public health. Globally, we are constantly trying to combat and prevent viral infection. With viruses everywhere, constantly undergoing reassortment and rapid evolution, it is important to try to understand and predict the next outbreak. Mode of transmission should be taken into account with degree of severity, as the right combination can pose the highest threat to humans: an easily transmitted fatal infection, as a strain could spontaneously emerge that render our immune systems and modern medications defenseless.
Background
The influenza viruses are RNA viruses which are classified as genera of the family Orthomyxoviridae (Merck, 2012). The three genera include influenza A, B and C. A distant relative of the parainfluenza viruses belonging to the paramyxovirus family, influenza most commonly affects the respiratory tracts of people of all ages (Merck, 2012).
Amongst all three genera, Influenza A is known to cause the most severe symptoms and be the most pathogenic to humans (Kawaoka, 2006). Influenza A originated from the natural hosts of wild aquatic birds (Kawaoka, 2006). The disease was transmitted from birds to humans through ingesting poultry. The H5N1 strain is the most well known avian strain, known as the "highly pathogenic avian influenza virus" (HPAI). It has been classified as a threat to humans and other animals, or panzootic. Most notable serotypes of influenza A which have been known to cause pandemics include: H1N1 (1918 Spanish flu and 2009 Swine flu), H2N2 (1957 Avian flu in Asia)
Wild aquatic birds are the natural hosts (Kawaoka, 2006). When transmitted from one species to another, for example, from birds to humans, the outbreaks can be disastrous, causing human pandemics or problems in domestic poultry. Some of the serotypes which are known to have caused the most severe pandemics are: H1N1 which was the cause of the Spanish flu in 1918 and Swine flu in 2009, H2N2 which was the source of the Avian flu in 1957 which was widespread in Asia (Kawaoka, 2006). H3N2 was the cause of the Hong Kong flu in 1968 and H5N1 which caused the Bird flu most recently in 2004 (Kawaoka, 2006).
Influenzavirus B is most often known to be pathogenic amongst humans, seals and ferrets but is much less common than influenzavirus A (Hay et al., 2001). A key reason why influenzavirus B exhibits less infectivity than influenzavirus A is due to its lack of mutation, as its mutation rate is about three times less than that of type A; this reduces the amount of heterogeneity of the virus (Hay et al., 2001). Immune systems are therefore able to combat and produce effective defenses against type B, such as antibodies, from an early age that withstand throughout lifetime (Hay et al., 2001). Influenzavirus C virus infects humans, dogs, and pigs. It is much less common than types A and B. It is most often seen in young children, as immune systems are not yet fully developed (Hay et al., 2001).
Currently, two FDA-approved antiviral medications exist known as Oseltamivir (Tamiflu) and Zanamivir. Oseltamivir and Zanamivir both utilize the same chemical mechanisms in order to prevent influenza spread. By inhibiting neuraminidase (NA), they are both affective against types A and B inhibiting the viral assembly process. Reported resistance to these treatments has thus so far been low. However, researchers are aware that this number could change easily increase, and are therefore trying to predict new mutations that could arise to help in search of new mechanisms for treatment. Resistance has also been reported to occur throughout treatment and even well after the final course of treatment.
Structure, properties, and subtype nomenclature
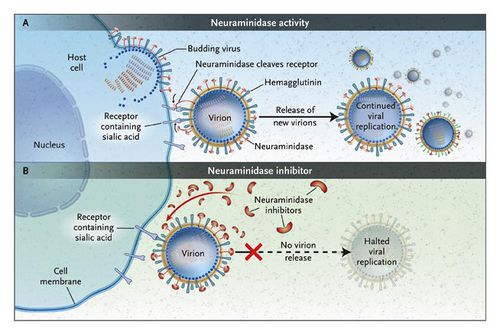
The size of the influenza particle is about 80-120 nanometers in diameter (NIH, 2005). The structure is typically sphere-like however variations due to filamentous structures can form, most commonly observed in type C (NIH, 2005). There is a viral envelope which encases the central core, housing and protecting the RNA genome. The envelope is made up of two types of glycoproteins, HA, hemagglutinin, and NA, neuraminidase. HA is a lectin or a carbohydrate protein whose role is to mediate the binding and entry processes of RNA into the host target cells. Alternatively, NA is more involved in the release of the progeny assembled viruses from host infected cells. Mechanistically, NA cleaves the sugars which keep the mature viral particles attached to the host cell. The HA and NA proteins are responsible for the type of antibody responses in the host cell and are also easy targets for inhibition of the virus used in antiviral drugs. Influenza A viruses are classified into serotypes based on the forms of HA and NA and ultimately their antibody responses, for example H1N1. There are 16 different HA and 9 different NA.
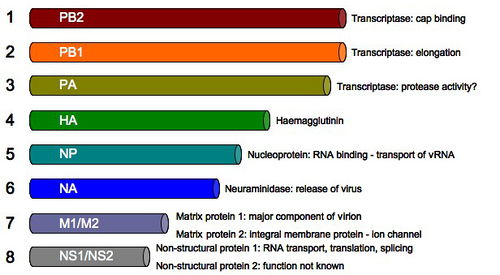
The genome of the influenza virus consists of 8 pieces of segmented negative sense RNA. Two strands encoding for the surface proteins: hemagglutinin (HA), neuraminidase (NA). PB2, the polymerase basic 2, encodes for the transcriptase involved in cap binding while PB1, the polymerase basic 1, is responsible for the transcriptase that facilitates elongation. The PA encodes for a transcriptase for protease activity. The NP, nucleoprotein, is involved in the RNA binding and transport. M1/M2, or the matrix proteins 1 and 2, are involved in producing the vital proteins in influenza such as ion channels. The NS1 is involved in RNA transport, translation and splicing while the NS2 encodes for unknown functions (Russell et al., 2012).
The sizes of the strands also vary in length. In descending order the strand size is: PB2, PB1, PA, HA, NP, NA, M1/M2 and NS1/NS2. For a virion to be complete, it must package all 8 RNA strand components. Therefore, there are many viruses that are assembled which are not viable.
Replication
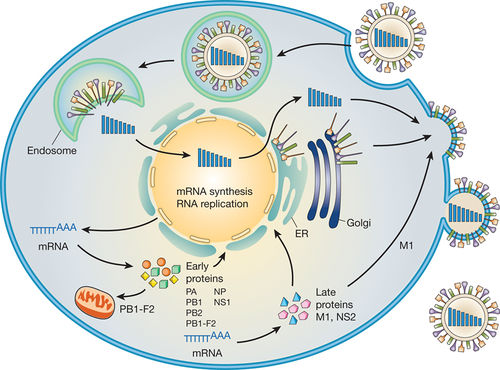
The influenza virus, like all viruses, replicate within their host cells; the process begins as the virus binds to the cell through the HA structure and receptors (Smith et al., 2005). Influenza binding occurs through HA onto sialic acid sugars on the surface of epithelial cells, typically found in mammals within the nose, throat and lungs, along with the intestines of birds (Smith et al., 2005). The HA is cleaved by a protease and the virus is endocytosed into the cell (Smith et al., 2005). As the virus binds and enters, the genome is transferred to a new host site where it can produce new copies of both its vital viral proteins for infectivity and RNA and essentially assemble new viral particles ultimately, budding from the cell (Smith et al., 2005). More specifically, the intracellular process begins as the virions come together into the microtubule organizing center and begin interaction with endosomes (Smith et al., 2005). The acidic conditions in the endosome allow for the HA proteins on the viral envelope to fuse with the vacuole’s membrane (Smith et al., 2005). Consequently, the M2 ion channel allows protons to penetrate the viral envelope and eventually acidify the core of the virus; this causes the virus to dissemble, releasing the viral RNA and core proteins (Smith et al., 2005). In amantadine drugs, the M2 ion channel is blocked, disrupting any infection (Smith et al., 2005). In the second stage of infection, the viral RNA (vRNA) molecules, accessory proteins and RNA-dependent RNA polymerase freely enter into the cytoplasm (Smith et al., 2005). The core proteins and vRNA come together to form a complex that is transported to the nucleus. Within the nucleus is where where RNA-dependent RNA polymerase transcribes a complementary positive sense vRNA; this vRNA can then follow either of two pathways, as it can be exported to the cytoplasm and translated there or can remain in the nucleus (Smith et al., 2005).
The newly synthesized viral proteins can be secreted through the Golgi apparatus onto the cell surface, i.e. the NA and HA surface proteins (Smith et al., 2005); proteins can also be transported back to the nucleus to produce new viral genome products through the vRNA (Smith et al., 2005). Other essential viral protein functions include: degrading the cellular mRNA and utilizing nucleotides for vRNA synthesis while prohibiting translation of host cell mRNA.
The final steps in the assembly of the virion include synthesis of the negative sense vRNAs, which alone form the genomes of future viruses, as well as the RNA-dependent RNA polymerase and all other viral proteins (Smith et al., 2005). The HA and NA molecules then come together in the host cell membrane (Smith et al., 2005). The vRNA along with the viral core proteins exit the nucleus and enter the same membrane area as HA and NA through a protrusion (Smith et al., 2005). The mature viruses then bud from the cell, encased in a sphere of phospholipid membrane, obtaining the HA and NA in the surface membrane layer (Smith et al., 2005). The viruses stay attached to the cell through HA and the mature viruses finally separates as the NA cleaves sialic acid residues from the host. After release of the fully formed viruses, the host ultimately dies (Smith et al., 2005).
The high mutation rate of the influenza virus which is the cause of the emergence of new strains leading to new infections, is largely due to the absence of an RNA proofreading enzyme (Drake et al., 1993). This absence causes the RNA-dependent RNA polymerase that copies the viral genome to make an error every 10 thousand nucleotides, about the length of the vRNA (Drake et al., 1993). A large majority of produced influenza viruses are typically mutants which contributes to the antigenic drift, or a change of the antigens on the viral surface, over time (Drake et al., 1993). The separation of the genome into eight distinct segments of vRNA also allows for re-assortment of vRNAs when more than one type of influenza virus infects a single cell (Drake et al., 1993). This mixing and re-assortment of vRNA allows for antigenic shift which is defined as the change from one antigen to another (Drake et al., 1993).
Transmission
Influenza has three main modes of transmission: direct, indirect and airborne. Direct transmission occurs when infected mucus through nose, eyes or throat, is passed into those of others. This involves direct contact of mucus membranes. Airborne transmission is produced through the respiratory tract, as aersols of an infected person are inhaled by a non-infected person. Liquid droplets small enough for inhalation are about 0.5-5μm in diameter. On average, one sneeze can produce up to 40,000 droplets, the majority of which are too big for airborne transmission to occur. The final mode of transmission is through indirect transmission, as transmission occurs through surface contact. Examples of common indirect contact include: skin to skin and skin to contaminated surface. Influenza can survive from up to 24-48 hours on sturdy non-porous surfaces. However, on paper dry surfaces, it can only survive up to 15 minutes. On skin, the virus can last up to 5 minutes. In mucus, the viruses are viable up to 17 days, making direct contact with mucosal membranes the most dangerous source of infection. Humidity and UV radiation are significant environmental factors that contribute to the transmission frequency as low humidity and lack of sunlight in the winter facilitate viral survival.
The shedding time of the influenza virus, or the duration of time in which an infected person might be infectious to others, begins about 24 hours before symptoms become overt (Hall et al., 2007). The virus is released between 5 to 7 days, however cases may vary. Peak of infection is most often observed between day 2 and 3 (Hall et al., 2007). Shedding time of the virus is also correlated with temperature of the host environment as hosts with fevers usually have higher transmission rates (Weber et al., 2008). Children are also much more infectious than adults, as shedding time can last up to two weeks after initial infection (Weber et al., 2008). These are all important elements when taking into consideration how influenza might affect specific as well as general populations at a given time; mathematical models have also been generated to predict effects (Weber et al., 2008).
Current Research
In one study conducted by Herfst et al. (2012) researchers sought to obtain an airborne transmissible A/H5N1 virus through ferrets. First researchers wanted to obtain a human transmissible virus and did so through selecting a mutant A/H5N1 virus with receptor specificity for α -2, 6-linked SA shed at high titers from the URT of ferrets. The specifity for α -2, 6- marks a shift from avian α -2, 3-linked SA receptors to human.
This was conducted through a QuickChange multisite-directed mutagenesis kit to introduce amino acid substitutions N182K/Q222L/G224S in the HA of wild-type (WT) A/Indonesia/5/2005, resulting in A/H5N1HA N182K , A/H5N1 HA Q222L, G224S and A/H5N1 HA N182K, Q222L, G224S
These experiment 1 three mutant viruses and A/H5N1 WT were inoculated into six ferrets for each group intra-nasally. Throat and nasal swabs were collected daily and virus titers were determined by end-point dilution in Madin Darby Canine Kidney (MDCK) cells to quantify virus shedding from the ferret URT. Three animals were euthanized after 3 days and the rest following after day 7; sample tissues were then taken. Virus titers were determined in the nasal turbinates, trachea and lungs collected post-mortem from the euthanized ferrets. Throughout the experiment, WT inoculated viruses produced up to 10 times more titers than A/H5N1 HA Q222L, G224S. Alternatively, A/H5N1 HA Q222L, G224S , produced the highest titers of all three mutants after 7 days. Notably, no significant difference was observed in virus shedding mutants within the first three days between A/H5N1 HA Q222L, G224S and A/H5N1 HA N182K when six animals were sampled in each group. Specificity for α-2,6-linked SA A/H5N1 HA Q222L,G224S yielded the highest virus titers in the ferret.
For the next experiment in the study, an additional amino acid substitution in E627K in PB2 in A/Indonesia/5/2005 was introduced through site mutagenesis, producing a recombinant A/H5N1 HA Q222L, G224S E627K. This substitution is known to be one of the most consistent host determinants in the virus. There was not a significant difference in virus shedding from this amino acid substitution. Both producing up to 1 x 104 50% tissue culture infectious doses (TCID50). Four ferrets were then housed into cages adjacent to those with four inoculated ferrets to test for airborne transmission. The A/H5N1 HA Q222L, G224S E627K was not found to be airborne transmissible. To force selection for a respiratory tract mammalian virus with the E627K mutation, one ferret was inoculated intra-nasally with WT and one with A/H5N1 HA Q222L, G224S E627K.. Throat and nose swabs were collected daily from live animals until 4 days post-inoculation (dpi) at which time animals were euthanized to collect samples from nasal turbinates and lungs. Nasal turbinates were homogenized in 3ml of virus transport medium, tissue debris was pelleted by centrifugation and 0.5 ml of supernatant was subsequently used to inoculate the next ferret intra-nasally with passage 2. Procedure was done repeatedly until passage 6. At passage 6, 1m of phosphate buffered saline (PBS) was delivered through drops into nostrils of ferrets to produce sneezing. About 200μl of the sneeze was collected and added to 2ml of PBS. Nasal wash samples were used for intranasal inoculation of ferrets for the passages of 7 through 10. Ten passages were conducted to ensure sufficient adaptation of the virus in mammals.
WT viruses turbinates ranged from 1x105 to 1x107 TCID50/gram tissue throughout the course of 10 serial passages. The ferrets inoculated with A/H5N1 HA Q222L, G224S E627K showed a slight increase in observed virus titers in nasal turbinates as passage number increased. The titer range was spread from 1x104 TCID50/gram tissue at the beginning of the experiment to about 3.2 x 105 to 1x106 TCID50/gram tissue in the final passages. Additionally, the nose swabs collected also increased throughout with peak shedding at passage 10. This suggests that A/H5N1 HA Q222L, G224S E627K had a greater amounts of replication in the inoculated ferret URT after repeated passage, adaption becoming more significantly apparent after passage 4.
Genetic composition of the viral quasi-species, also known as the mixture of viruses with a variety of mutations, was found in the nasal washes after 10 passages of both of the WT and A/H5N1 HA Q222L, G224S E627K . This was determined by sequence analysis using the 545/Roche GS-FLX sequencing platform. Mutations in the A/H5N1 HA Q222L, G224S E627K remained present in population after 10 consecutive passages with a frequency greater than 99.5%. No mutations were found in either virus types in segment 7. The only amino acid substitution detected upon repeated passage in both WT and A/H5N1 HA Q222L, G224S E627K was T156A in HA, removing a potential N-linked glycosylation site in HA, detected in 99.6% of viruses in WT after 10 passages and in 89% of the A/H5N1 HA Q222L, G224S E627K . The remaining 11% showed substitutions in the N154K which removes the same potential N-linked glycosylation in site HA. Essentially in all viruses past passage 10, this mutation in the HA site was present.
In their final experiment researchers in this study wanted to test for airborne transmissible viruses present after passage 10 in the ferrets for both WT and A/H5N1 HA Q222L, G224S E627K. They did this through nasal washes which were collected at 3 dpi from ferrets at passage 10. Nasal Wash samples were diluted 1:2 in PBS and next used to inoculate into six ferrets intranasally two with WT and four with A/H5N1 HA Q222L, G224S E627K . Naïve recipient ferret was placed in cage adjacent to inoculated donor ferret. Experimental set-up includes cages that allow direct airflow but no contact betwen a donor ferret into neighboring recipient. Naïve swabs were then collected and inoculated in MDCK cells. There was no replicating virus in MDCK cells with WT, however there was with A/H5N1 HA Q222L, G224S E627K. Three of the four naïve recipients in the A/H5N1 HA Q222L, G224S E627K became infected. Ferret 2 of the airborne infected A/H5N1 HA Q222L, G224S E627K contained highest amount of virus titer among ferrets and was used to inoculate and create airborne transmissible version with 6 other ferrets.
The genome sequences of viruses with airborne transmission all had Q22L, G224S and E627K mutations, only two additional amino acid substitutions both in HA was detected in all six airborne viruses. H103Y which forms part of the HA trimer interface and T156A which is proximal to the RBS. There were also many other mutations but none of which were consistent throughout heterogeneous virus populations
Overall, results have allowed for researchers to hypothesize that five amino acid substitutions, four in HA and one in PB2 may be sufficient to confer airborne transmission of HPAI A/H5N1 virus between mammals. This causes a potential threat to humans and the next pandemic of influenza. Through mutations from co-infecting viruses or through random evolution, we become more susceptible to a highly airborne transmittable virus with severe pathogenic characteristics, or the next pandemic.
References
Drake, J. 1993. Rates of spontaneous mutation among RNA viruses" Proc Natl Acad Sci USA 90 (9): 4171–5
Hall CB. 2007. The spread of influenza and other respiratory viruses: complexities and conjectures. Clin. Infect. Dis. 45 (3): 353–9.
Hay, A; Gregory V, Douglas A, Lin Y. 2001. The evolution of human influenza viruses. Philos Trans R Soc Lond B Biol Sci 356 (1416): 1861–70 Herfest , Sander et al. 2012. Airborne Transmission of Influenza A/H5N1 Virus Between Ferrets. Science 336, 1534.
Kawaoka Y. 2006. Influenza Virology: Current Topics. Caister Academic Press. Merck, Eccles, R. 2005. Understanding the symptoms of the common cold and influenza. Influenza: Viral Infections: Merck Manual Home Edition. Lancet Infect Dis 5 (11): 718–25.
Russell, Colin A, Fonville, Judith M; Brown, André E X, David F Burke; David L Smith; Sarah L James; Sander Herfst; Sander van Boheemen; Martin Linster; Eefje J Schrauwen; et al. 2012. The potential for respiratory droplet-transmissible A/H5N1 influenza virus to evolve in a mammalian host. Department of Zoology, University of Cambridge, Cambridge, UK. Science (New York, N.Y.)
Smith AE, Helenius A. 2004. How viruses enter animal cells. Science 304 (5668): 237–42.
Shindo, Nikki Dr. Transcript of virtual press conference with Gregory Hartl, Spokesperson for H1N1, and Dr Nikki Shindo, Medical Officer, Global Influenza Programme, World Health Organization. 2009. World Health Organization.
Vainionpää R, Hyypiä T. 1994. Biology of parainfluenza viruses. Clin. Microbiol. Rev. 7 (2): 265–75.
International Committee on Taxonomy of Viruses descriptions of:Orthomyxoviridae, Influenzavirus Band Influenzavirus C. National Institutue of Health. 2005.
Weber TP, Stilianakis NI. 2008. Inactivation of influenza A viruses in the environment and modes of transmission: a critical review. J. Infect. 57 (5): 361–73.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2011, Kenyon College.