User:Rachelevelyn7: Difference between revisions
No edit summary |
mNo edit summary |
||
| Line 28: | Line 28: | ||
In many agricultural areas in North America the nitrate concentrations exceed the standards. [http://en.wikipedia.org/wiki/Denitrifying_bacteria Denitrifying organisms] are capable of using nitrate or nitrite as terminal electron acceptors thereby removing the excess of nitrogen from the environment. The organism Methylomirabilis oxyfera is an example of such an organism. This denitrifying bacterium is special in that it doesn’t have the gene encoding nitrous oxide reductase, the protein that converts N2O to N2. Instead they harbor an operon which encodes the complete methane monooxygenase complex. This enables it to oxide methane in an aerobic pathway [[#References|[1]]]. The mechanism takes advances of the oxidation of methane to drive denitrification. They do so by producing oxygen from nitrite via nitrite oxide (thereby bypassing the intermediate nitrous oxide) and then use this oxygen to oxide methane in an anaerobic environment. This is called nitrite dependent anaerobic methane oxidation. The overall redox reaction is 3CH4 + 8NO2- + 8H+ -> 3CO2 + 4N2 + 10 H2O. In this way the organism uses the potent greenhouse gas methane and reduces nitrite thereby contributing to the removal of excess N-compounds in groundwater [[#References|[1]]]. | In many agricultural areas in North America the nitrate concentrations exceed the standards. [http://en.wikipedia.org/wiki/Denitrifying_bacteria Denitrifying organisms] are capable of using nitrate or nitrite as terminal electron acceptors thereby removing the excess of nitrogen from the environment. The organism Methylomirabilis oxyfera is an example of such an organism. This denitrifying bacterium is special in that it doesn’t have the gene encoding nitrous oxide reductase, the protein that converts N2O to N2. Instead they harbor an operon which encodes the complete methane monooxygenase complex. This enables it to oxide methane in an aerobic pathway [[#References|[1]]]. The mechanism takes advances of the oxidation of methane to drive denitrification. They do so by producing oxygen from nitrite via nitrite oxide (thereby bypassing the intermediate nitrous oxide) and then use this oxygen to oxide methane in an anaerobic environment. This is called nitrite dependent anaerobic methane oxidation. The overall redox reaction is 3CH4 + 8NO2- + 8H+ -> 3CO2 + 4N2 + 10 H2O. In this way the organism uses the potent greenhouse gas methane and reduces nitrite thereby contributing to the removal of excess N-compounds in groundwater [[#References|[1]]]. | ||
[[File:Pumpandtreat.gif|thumb|400px|right|]] | [[File:Pumpandtreat.gif|thumb|400px|right|]] | ||
| Line 41: | Line 40: | ||
Several components of the rhizobium-legume interactions are similar to | Several components of the rhizobium-legume interactions are similar to | ||
[http://en.wikipedia.org/wiki/Arbuscular_mycorrhiza arbuscular mycorrhizal | [http://en.wikipedia.org/wiki/Arbuscular_mycorrhiza arbuscular mycorrhizal | ||
(AM) symbiosis]. In both of these symbioses, specialized host membranes surround the microbes and form a symbiotic surface that facilitates the exchange of nutrients [1]. Another similarity is that AM fungi enter the root of their host plant in a comparable mechanism to rhizobia [16]. Also, the [http://en.wikipedia.org/wiki/Hypha hyphae] of the AM fungi spread within the cells of the inner cortex of the plant, this is mimicked by the rhizobium infection threads when forming nodules within the inner cortex of legumes [15]. Significantly, AM fungi produce lipochito-oligosaccharides, which are structurally similar to the Nod factors synthesized by rhizobia [15]. | (AM) symbiosis]. In both of these symbioses, specialized host membranes surround the microbes and form a symbiotic surface that facilitates the exchange of nutrients [[#References|[1]]]. Another similarity is that AM fungi enter the root of their host plant in a comparable mechanism to rhizobia [[#References|[16]]]. Also, the [http://en.wikipedia.org/wiki/Hypha hyphae] of the AM fungi spread within the cells of the inner cortex of the plant, this is mimicked by the rhizobium infection threads when forming nodules within the inner cortex of legumes [[#References|[15]]]. Significantly, AM fungi produce lipochito-oligosaccharides, which are structurally similar to the Nod factors synthesized by rhizobia [[#References|[15]]]. | ||
Thus, it has been hypothesized that these similar cellular processes are evidence that rhizobium-legume interactions was derived from the AM symbiosis with the roots of plants and the similar interactions between these two symbioses have been compiled into a common signalling pathway [16]. | Thus, it has been hypothesized that these similar cellular processes are evidence that rhizobium-legume interactions was derived from the AM symbiosis with the roots of plants and the similar interactions between these two symbioses have been compiled into a common signalling pathway [[#References|[16]]]. | ||
=References= | =References= | ||
Revision as of 15:20, 13 December 2012
I am a third year science undergrad at the University of British Columbia. I am working towards a Bachelor's in Microbiology and Immunology.
Hello :)
Nod Factors
Introduction
Groundwater serves as water for more than 50% of people living in North America therefore a significant public resource. To date, major contamination of groundwater in North America are due to the release and use of chlorinated ethenes by industry. Examples of such toxic compounds are perchloroethene (PCE), trichloroethene (TCE). Carbon tetrachloride (CT) is also a major groundwater pollutant [4]. These compounds were widely used as solvents for dry cleaning and in textile manufacturing. They are sufficiently water soluble and can travel through soil where they reach the groundwater. The relative high concentration of them here can be harmful [6]. Ground water is also contaminated by pollutants that are not highly toxic, but can be utilized or modified by microorganisms to become more toxic. For instance over-fertilization in agriculture leads to an increased nitrate concentration which i.e. can cause the Blue Baby syndrome. This is seen in infants younger than six month old who rely on bacteria to digest their food. Some of these bacteria also convert nitrate, a component of fertilizer, to nitrite. In the blood nitrite reacts with hemoglobin interfering with its ability to carry oxygen. The babies show sign of suffocation and gets a bluish skin [2].
Rhizobium-Legume Symbiosis
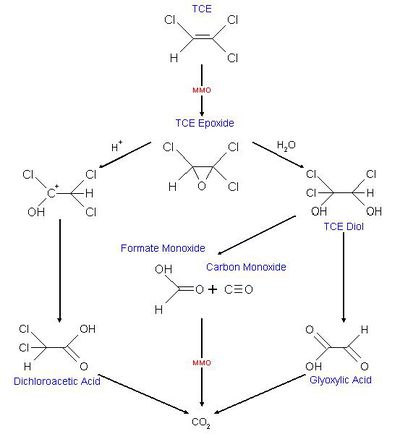
Some dehalorespiring organisms are capable of degrading PCE, TCE and CT into non-toxic compounds. Degradation of PCE is only known to happen through reductive dechlorination and only under anaerobic condition. TCE is, unlike PCE, able to be degraded under aerobic conditions. This can happen through cometabolism. In co-metabolism a compound is transformed by an organism that doesn’t use the compound as an energy or carbon source and reducing power is not provided. The organism relies on another compound to serve as an energy and carbon source [3] . Methanotrophic organisms grow on methane as a primary substrate and oxygen but some are also able to degrade TCE as a secondary substrate. This is because of nonspecific enzymatic activity of enzymes (methane monooxygenase, MMO) involved in degradation of the primary substrate. The degradation of TCE serves no beneficial purpose for these organisms. It generates an epoxide(cf. figure 1) which is transported out of the cell and here other heterotrophic organisms bring about the transformation into non-toxic compounds resulting in the formation of CO2. Several factors inhibit the aerobic degradation of TCE here among the concentration of contamination, the pH and the temperature. Because both TCE and methane bind to the same site in MMO competition between growth substrate and non-growth substrate also seems to limit degradation of TCE [3].
Nodulation Process
The bacterium Pseudomonas stutzeri strain KC can dehalogenate CT into carbon dioxide and chlorine without producing the toxic intermediate chloroform (CCl3H). This bacterium is originally isolated from an aquifer in Seal Beach in California. It is dependent on anaerobic conditions and in iron-limited media this bacterium produces and secretes a chelator called pyridine-2,6 (bis)thiocarboxylate (PDTC cf. figure 2.) [5]. When PDTC is in contact with a broad range of cell components it turns into a reduced form (the iron in the complex is reduced) and this is essential for its extracellular activity. PDCT has to be in a complex with copper in order for the fast turnover rate of CT into CO2. This complex functions both as a reactant and a catalyst in the reaction. When Pseudomonas stutzeri is in environments were nitrate is present as the electron acceptor a more rapid production of PDTC is observed [6].
Function of Nod Factors
In many agricultural areas in North America the nitrate concentrations exceed the standards. Denitrifying organisms are capable of using nitrate or nitrite as terminal electron acceptors thereby removing the excess of nitrogen from the environment. The organism Methylomirabilis oxyfera is an example of such an organism. This denitrifying bacterium is special in that it doesn’t have the gene encoding nitrous oxide reductase, the protein that converts N2O to N2. Instead they harbor an operon which encodes the complete methane monooxygenase complex. This enables it to oxide methane in an aerobic pathway [1]. The mechanism takes advances of the oxidation of methane to drive denitrification. They do so by producing oxygen from nitrite via nitrite oxide (thereby bypassing the intermediate nitrous oxide) and then use this oxygen to oxide methane in an anaerobic environment. This is called nitrite dependent anaerobic methane oxidation. The overall redox reaction is 3CH4 + 8NO2- + 8H+ -> 3CO2 + 4N2 + 10 H2O. In this way the organism uses the potent greenhouse gas methane and reduces nitrite thereby contributing to the removal of excess N-compounds in groundwater [1].
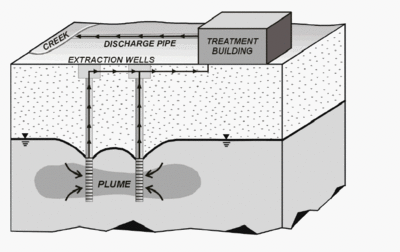
Contamination of groundwater can lead to severe health problems and environmental changes if left untreated. Bioaugmentation is a widespread biological technique used in the removal of chlorinated compounds. By introducing natural electron donors that are helpful in the removal of halogenated compounds into the groundwater the growth of dehalorespiring organisms can be favored. Optional conditions for dehalogenation are provided without any engineering steps taken [7]. Pump and treat method is also one of the most used groundwater remediation techniques. Removal of contaminated groundwater from soil with the use of pumps followed by subsequent remediation at the surface helps overcome the persistence of the pollutants (cf. figure 3). It is typically biological or chemical treatments that remove the pollutants. This method is costly and slow however and some contaminants cannot be removed because they stick to soil and rocks or are not sufficient water soluble [4].
Host Specificity due to Nod Factors
Genetic History
Several components of the rhizobium-legume interactions are similar to [http://en.wikipedia.org/wiki/Arbuscular_mycorrhiza arbuscular mycorrhizal (AM) symbiosis]. In both of these symbioses, specialized host membranes surround the microbes and form a symbiotic surface that facilitates the exchange of nutrients [1]. Another similarity is that AM fungi enter the root of their host plant in a comparable mechanism to rhizobia [16]. Also, the hyphae of the AM fungi spread within the cells of the inner cortex of the plant, this is mimicked by the rhizobium infection threads when forming nodules within the inner cortex of legumes [15]. Significantly, AM fungi produce lipochito-oligosaccharides, which are structurally similar to the Nod factors synthesized by rhizobia [15]. Thus, it has been hypothesized that these similar cellular processes are evidence that rhizobium-legume interactions was derived from the AM symbiosis with the roots of plants and the similar interactions between these two symbioses have been compiled into a common signalling pathway [16].
References
(1) Catoira, R., Galera, C. de Billy, F., Penmetsa, R. V., Journet, E.-P., Maillet, F., Rosenberg, C., Cook, D., Gough, C., and Denarie, J. “Four Genes of Medicago truncatula Controlling Components of a Nod Factor Transduction Pathway.” 2000. The Plant Cell, Volume 12. p. 1647-1665.
(2) Dakora, F. D. “Defining New Roles for Plant and Rhizobial Molecules in Sole and Mixed Plant Cultures Involving Symbiotic Legumes.” 2003. New Phytologist, Volume 158. p. 39-49.
(3) Guerts, R., Lillo, A., and Bisseling, T. “Exploiting an ancient signalling machinery to enjoy a nitrogen fixing symbiosis.” 2012. Current Opinion in Plant Biology. Volume 15. p. 438-443.
(4) Postgate, J. (1998). Nitrogen Fixation, 3rd Edition. Cambridge University Press. Cambridge UK.
(5) Zumft, W. G. and Mortenson, L. E. “The nitrogen-fixing complex of bacteria.” 1975. Biochem. Biophys. Acta. Volume 416. p. 1-52.
(6) Akcay, E. and Roughgarden, J. “Negotiation of Mutualism: Rhizobia and Legumes.” 2007. Proceedings: Biological Sciences, Volume 274. p. 25-32.
(7) Depret, G. and Laguerre, G. “Plant Phenology and Genetic Variability in Root and Nodule Development Strongly Influence Genetic Structure of Rhizobium Leguminosarum Biovar Viciae Populations Nodulating Pea.” 2008. New Phytologist, Volume 179. p. 224-235.
(8) Lindstrom, K., Terefework, Z., Suominen, L., and Lortet, G. “Signalling and Development of Rhizobium: Legume Symbioses.” 2002. Biology and Environment: Proceedings of the Royal Irish Academy, Volume 102B. p. 61-64.
(9) Lodwig, E. M., Hosie, A. H., Bourdes, A., Findlay, K., Allaway, D., Karunakaran, R., Downie, J. A., and Poole, P. S. “Amino-acid cycling drives nitrogen fixation in the legume-Rhizobium symbiosis.” 2003. Nature, Volume 422. p. 722-726.
(10) Streng, A., Camp, R., Bisseling, T., and Guerts, R. “Evolutionary origin of rhizobium Nod factor signaling.” 2011. Plant Signaling and Behaviour, Volume 6. p. 1510-1514
(11) Schultze, M., Quiclet-Sire, B., Kondorosi, E., Virelizier, H., Glushak, J. N., Endre, G., Gero, S. D., and Kondorosi, A. “Rhizobium meliloti Produces a Family of Sulfated Lipo-Oligosaccharides Exhibiting Different Degrees of Plant Host Specificity. 1992. Proceedings of the National Academy of Sciences of the United States of America, Volume 89. p. 192-196.
(12) Perret, X., Staehelin, C., and Broughton, W. J., “Molecular Basis of Symbiotic Promiscuity.” 2000. Microbiology and Molecular Biology Reviews, Volume 64. p. 180-201.
(13) Ardourel, M., Demont, N., Debelle, F., Maillet, F., de Billy, F., Prome, J-C., Denarie, J., and Truchet, G. “Rhizobium meliloti Lipooligosaccharide Nodulation Factors: Different Structural Requirements for Bacterial Entry into Target Root Hair Cells and Induction of Plant Symbiotic Developmental Responses.” 1994. The Plant Cell, Volume 6. p. 1357-1374.
(14) Vijn, I., das Neves, L., van Kammen, A., Franssen, H., and Bissling, T. “Nod Factors and Nodulation in Plants.” 1993. Science, Volume 260. p. 1764-1765.
(15) Bonfante, P. and Requena, N. “Dating in the dark: how roots respond to fungal signals to establish arbuscular mycorrhizal symbiosis.” 2011. Current Opinion in Plant Biology, Volume 14. p. 451-457.
(16) Ivanov, S., Fedorova, E. E., Limpens, E., De Mita, S., Genre, A., Bonfante, P., and Bisseling, T. “Rhizobium-legume symbiosis shares an exocytotic pathway required for arbuscule formation. 2012. Proceedings of the National Academy of Sciences of the United States of America, Volume 109. p. 8316-8321.