User:S4342592: Difference between revisions
No edit summary |
|||
| Line 17: | Line 17: | ||
''Porphyromonas gingivalis'' strain 2561 = ATCC 33277= CCUG 25893 = CCUG 25928 = CIP 103683 = DSM 20709 = JCM 12257 = NCTC 11834. | ''Porphyromonas gingivalis'' strain 2561 = ATCC 33277= CCUG 25893 = CCUG 25928 = CIP 103683 = DSM 20709 = JCM 12257 = NCTC 11834. | ||
[[File:blackcolonies.jpeg|thumb|Black pigmentation of ''Porphyromonas gingivalis'' on blood agar <sup>[[#References|[ | [[File:blackcolonies.jpeg|thumb|Black pigmentation of ''Porphyromonas gingivalis'' on blood agar <sup>[[#References|[2]]]</sup>]] | ||
==Description and significance== | ==Description and significance== | ||
''Porphyromonas gingivalis'' is a gram negative, non motile, rod shaped bacteria that is pathogenic, with no functional role <sup>[[#References|[1]]]</sup>. It is commonly found in the oral cavity | ''Porphyromonas gingivalis'' is a gram negative, non motile, rod shaped bacteria that is pathogenic, with no functional role <sup>[[#References|[1]]]</sup>. It is commonly found in the oral cavity. On blood agar plate, it forms black pigmented colonies due to the build-up of hemin that helps to transport iron <sup>[[#References|[2]]]</sup>. ''P. gingivalis'' is the main source of periodontal disease and is also said to be associated with cardiovascular disease and diabetes amongst other inflammatory disease thus it is important to study this bacterium, to help reduce the risk of more serious illnesses <sup>[[#References|[3]]]</sup>. | ||
==Genome structure== | ==Genome structure== | ||
In 2003, the genome of ''P. gingivalis'' was described to have 2,343,479bp with an average guanine cytosine nucleotide content of 48.3%. It has 4 ribosomal operons, 2 RNA structural genes and 53 tRNA gene. There was a total of 1990 open reading frames that could be identified in the genome. 21 areas of the genome were discovered to display an atypical nucleotide composition. The areas range in size from 11 to 68kb and has a guanine cytosine nucleotide content of between 29.4% to 61.6%. 463 genes were also seen in the genome <sup>[[#References|[ | In 2003, the genome of ''P. gingivalis'' was described to have 2,343,479bp with an average guanine cytosine nucleotide content of 48.3%. It has 4 ribosomal operons, 2 RNA structural genes and 53 tRNA gene. There was a total of 1990 open reading frames that could be identified in the genome. 21 areas of the genome were discovered to display an atypical nucleotide composition. The areas range in size from 11 to 68kb and has a guanine cytosine nucleotide content of between 29.4% to 61.6%. 463 genes were also seen in the genome <sup>[[#References|[4]]]</sup><sup>[[#References|[5]]]</sup> | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
| Line 35: | Line 32: | ||
Fimbriae are thin, proteinaceous surface appendages that protrudes from the outer membrane of a bacterial cell. Its function is essential for invasion of host cells, adherence to oral substrates as well as contribution to the progression of periodontal inflammatory reactions. | Fimbriae are thin, proteinaceous surface appendages that protrudes from the outer membrane of a bacterial cell. Its function is essential for invasion of host cells, adherence to oral substrates as well as contribution to the progression of periodontal inflammatory reactions. | ||
Lipopolysaccharide (LPS) is also an important component of the bacterial outer membrane. It helps to facilitate bacterial survival and maintain the cellular and structural integrity as well as controlling the entry of hydrophobic molecules and toxic chemicals. | Lipopolysaccharide (LPS) is also an important component of the bacterial outer membrane. It helps to facilitate bacterial survival and maintain the cellular and structural integrity as well as controlling the entry of hydrophobic molecules and toxic chemicals. | ||
The outer membrane that contains the fimbriae and LPS amongst other structures, is an asymmetrical bilayer that plays in role as a selective barrier that offers protection and allow movement of various substrate though the outer membrane porin proteins <sup>[[#References|[]]]</sup>. | The outer membrane that contains the fimbriae and LPS amongst other structures, is an asymmetrical bilayer that plays in role as a selective barrier that offers protection and allow movement of various substrate though the outer membrane porin proteins <sup>[[#References|[6]]]</sup>. | ||
===biofilm formation=== | ===biofilm formation=== | ||
Dental plaque develops on tooth surfaces and in certain circumstances it progresses to a more pathogenic condition such as periodontal diseases. The process of plaque build-up is characterised by binding interactions of various oral bacteria. Early, primary commensal colonisers such as oral streptococci attach to the enamel pellicle, covering the tooth surface and providing attachment sites for subsequent colonisation by other potential pathogenic bacteria such as ''P. gingivalis''. Evidently, together with ''Streptococcus gordonii'', ''P. gingivalis'' is able to colonise efficiently, making it an important process in biofilm formation <sup>[[#References|[]]]</sup>. | Dental plaque develops on tooth surfaces and in certain circumstances it progresses to a more pathogenic condition such as periodontal diseases. The process of plaque build-up is characterised by binding interactions of various oral bacteria. Early, primary commensal colonisers such as oral streptococci attach to the enamel pellicle, covering the tooth surface and providing attachment sites for subsequent colonisation by other potential pathogenic bacteria such as ''P. gingivalis''. Evidently, together with ''Streptococcus gordonii'', ''P. gingivalis'' is able to colonise efficiently, making it an important process in biofilm formation <sup>[[#References|[7]]]</sup>. | ||
===Metabolic Functions=== | ===Metabolic Functions=== | ||
Genome analysis suggest various metabolic pathways that ''P. gingivalis'' is capable of. There are also some metabolic pathways that ''P. gingivalis'' perform poorly or not able to perform. ''P. gingivalis'' has a limited capability for organic nutrient metabolism and poor glucose utilisation. In addition, carbohydrates do not support the gorwth of the bacteria. However, ''P. gingivitis'' contain open reading frames (ORF) for all enzymes that is necessary for glycolytic pathway and ORF necessary for glucose/galactose transporters and glucose kinase. Pentose phosphate pathway was identified for anaerobic growth. Amino acid is also available for energy production. Other pathways that ''P. gingivalis'' is capable of performing include lysine catabolic pathway and iron acquisition <sup>[[#References|[]]]</sup>. | Genome analysis suggest various metabolic pathways that ''P. gingivalis'' is capable of. There are also some metabolic pathways that ''P. gingivalis'' perform poorly or not able to perform. ''P. gingivalis'' has a limited capability for organic nutrient metabolism and poor glucose utilisation. In addition, carbohydrates do not support the gorwth of the bacteria. However, ''P. gingivitis'' contain open reading frames (ORF) for all enzymes that is necessary for glycolytic pathway and ORF necessary for glucose/galactose transporters and glucose kinase. Pentose phosphate pathway was identified for anaerobic growth. Amino acid is also available for energy production. Other pathways that ''P. gingivalis'' is capable of performing include lysine catabolic pathway and iron acquisition <sup>[[#References|[8]]]</sup>. | ||
==Ecology== | ==Ecology== | ||
''P. gingivalis'' is an anaerobic rod shaped microorganism that is mainly found in the subgingival plaque of the periodontal in the oral cavity. Other potential environment where the pathogenic bacteria can be found include the upper gastrointestinal tract, the colon and the respiratory tract <sup>[[#References|[]]]</sup>. it has also been found in women with bacterial vaginosis <sup>[[#References|[]]]</sup> | ''P. gingivalis'' is an anaerobic rod shaped microorganism that is mainly found in the subgingival plaque of the periodontal in the oral cavity. Other potential environment where the pathogenic bacteria can be found include the upper gastrointestinal tract, the colon and the respiratory tract <sup>[[#References|[9]]]</sup>. it has also been found in women with bacterial vaginosis <sup>[[#References|[10]]]</sup> | ||
===Microbe/Host interactions=== | ===Microbe/Host interactions=== | ||
[[File:periodontaldisease.jpeg|thumb|300px|The state and progression of periodontal disease in comparison to a healthy gum. Periodontal disease is characterised by the teeth pockets and loss of bone mass of the teeth <sup>[[#References|[]]]</sup>]] | [[File:periodontaldisease.jpeg|thumb|300px|The state and progression of periodontal disease in comparison to a healthy gum. Periodontal disease is characterised by the teeth pockets and loss of bone mass of the teeth <sup>[[#References|[12]]]</sup>]] | ||
==Pathology== | ==Pathology== | ||
''P. gingivalis'' is responsible for causing periodontal disease. Periodontal disease is characterized by the destruction of tooth supporting tissues due to complex polymicrobial infection. The disease progresses from acute inflammation of the gingival tissue to formation of tooth pockets which eventually leads to the loss of teeth. Symptoms of periodontal disease include swollen or bleeding gums, sensitive teeth and receding gums | ''P. gingivalis'' is responsible for causing periodontal disease. Periodontal disease is characterized by the destruction of tooth supporting tissues due to complex polymicrobial infection. The disease progresses from acute inflammation of the gingival tissue to formation of tooth pockets which eventually leads to the loss of teeth. Symptoms of periodontal disease include swollen or bleeding gums, sensitive teeth and receding gums <sup>[[#References|[11]]]</sup>]] | ||
Other than causing periodontal disease, the microorganism also cause rheumatoid arthritis. | Other than causing periodontal disease, the microorganism also cause rheumatoid arthritis. <sup>[[#References|[13]]]</sup>]] | ||
==Application to biotechnology== | ==Application to biotechnology== | ||
Revision as of 08:39, 23 September 2016
Nur' Amirah Mohd Kassim Bench D 09082016
Classification
Higher order taxa
- Kingdom: Bacteria
- Domain: Bacteria
- Phylum: Bacteroidetes
- Class: Bacteroidia
- Order: Bacteroidales
- Family: Porphyromonadaceae
- Genus: Porphyromonas
Species
Porphyromonas gingivalis strain 2561 = ATCC 33277= CCUG 25893 = CCUG 25928 = CIP 103683 = DSM 20709 = JCM 12257 = NCTC 11834.

Description and significance
Porphyromonas gingivalis is a gram negative, non motile, rod shaped bacteria that is pathogenic, with no functional role [1]. It is commonly found in the oral cavity. On blood agar plate, it forms black pigmented colonies due to the build-up of hemin that helps to transport iron [2]. P. gingivalis is the main source of periodontal disease and is also said to be associated with cardiovascular disease and diabetes amongst other inflammatory disease thus it is important to study this bacterium, to help reduce the risk of more serious illnesses [3].
Genome structure
In 2003, the genome of P. gingivalis was described to have 2,343,479bp with an average guanine cytosine nucleotide content of 48.3%. It has 4 ribosomal operons, 2 RNA structural genes and 53 tRNA gene. There was a total of 1990 open reading frames that could be identified in the genome. 21 areas of the genome were discovered to display an atypical nucleotide composition. The areas range in size from 11 to 68kb and has a guanine cytosine nucleotide content of between 29.4% to 61.6%. 463 genes were also seen in the genome [4][5]
Cell structure and metabolism
Cell wall
To get establish in the oral cavity, microorganism have to adhere to the teeth surface. Adhesins are molecules found in the capsule of the cell wall that allows the adhesion. The capsule is therefore responsible in the initial adhesion to the human periodontal pocket epithelial cells, where it helps facilitate attachment. Fimbriae are thin, proteinaceous surface appendages that protrudes from the outer membrane of a bacterial cell. Its function is essential for invasion of host cells, adherence to oral substrates as well as contribution to the progression of periodontal inflammatory reactions. Lipopolysaccharide (LPS) is also an important component of the bacterial outer membrane. It helps to facilitate bacterial survival and maintain the cellular and structural integrity as well as controlling the entry of hydrophobic molecules and toxic chemicals. The outer membrane that contains the fimbriae and LPS amongst other structures, is an asymmetrical bilayer that plays in role as a selective barrier that offers protection and allow movement of various substrate though the outer membrane porin proteins [6].
biofilm formation
Dental plaque develops on tooth surfaces and in certain circumstances it progresses to a more pathogenic condition such as periodontal diseases. The process of plaque build-up is characterised by binding interactions of various oral bacteria. Early, primary commensal colonisers such as oral streptococci attach to the enamel pellicle, covering the tooth surface and providing attachment sites for subsequent colonisation by other potential pathogenic bacteria such as P. gingivalis. Evidently, together with Streptococcus gordonii, P. gingivalis is able to colonise efficiently, making it an important process in biofilm formation [7].
Metabolic Functions
Genome analysis suggest various metabolic pathways that P. gingivalis is capable of. There are also some metabolic pathways that P. gingivalis perform poorly or not able to perform. P. gingivalis has a limited capability for organic nutrient metabolism and poor glucose utilisation. In addition, carbohydrates do not support the gorwth of the bacteria. However, P. gingivitis contain open reading frames (ORF) for all enzymes that is necessary for glycolytic pathway and ORF necessary for glucose/galactose transporters and glucose kinase. Pentose phosphate pathway was identified for anaerobic growth. Amino acid is also available for energy production. Other pathways that P. gingivalis is capable of performing include lysine catabolic pathway and iron acquisition [8].
Ecology
P. gingivalis is an anaerobic rod shaped microorganism that is mainly found in the subgingival plaque of the periodontal in the oral cavity. Other potential environment where the pathogenic bacteria can be found include the upper gastrointestinal tract, the colon and the respiratory tract [9]. it has also been found in women with bacterial vaginosis [10]
Microbe/Host interactions
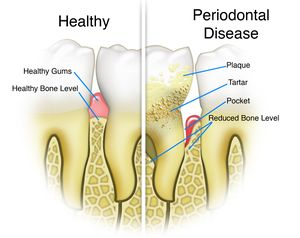
Pathology
P. gingivalis is responsible for causing periodontal disease. Periodontal disease is characterized by the destruction of tooth supporting tissues due to complex polymicrobial infection. The disease progresses from acute inflammation of the gingival tissue to formation of tooth pockets which eventually leads to the loss of teeth. Symptoms of periodontal disease include swollen or bleeding gums, sensitive teeth and receding gums [11]]]
Other than causing periodontal disease, the microorganism also cause rheumatoid arthritis. [13]]]
Application to biotechnology
Given the emerging evidence that periodontal infections and systemic conditions has been closely associated with one another, the search for a method to detect the presence of P. gingivalis is therefore necessary. Characterised antibodies has been raised against purified P. gingivalis HmuY protein and elected epitopes of the HmuY molecule. The results obtain from the study suggest that P. gingivalis HmuY protein may be used as an antigen for antibodies to target this bacteria. Therefore, purified HmuY protein will be use to suggest levels of anti-P. gingivalis antibodies in the sera of patients with chronic periodontitis []. In another study, various plant derived compounds such as quercetin and resveratol has shown inhibitory effects on the activity of P. gingivalis fimbriae, however, further studies needs to be conducted in order to finally produce a effective inhibitor against P. gingivalis [].
Current research
Summarise some of the most recent discoveries regarding this species.
References
References examples
This page is written by Nur' Amirah Mohd Kassim for the MICR3004 course, Semester 2, 2016
