Vibrio cholerae O1 Biotype El Tor: Virulence Factors: Difference between revisions
| Line 9: | Line 9: | ||
<i>Vibrio cholerae</i> serogroup O1 biotype El Tor (<i>V.cholerae</i> O1 El Tor) was the dominant strain of the 7th global cholera pandemic. It possesses two circular chromosomes that encode for 3,885 total open reading frames (2,770 on Chr 1 and 1,115 on Chr 2). <ref name=S2> [https://doi.org/10.1038/35020000 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.]</ref> Most essential cell function and pathogenicity genes – including those for DNA replication and toxin production – are on Chr 1. <ref name=S2> [https://doi.org/10.1038/35020000 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.]</ref> The ability of V. cholerae to cause illness in hosts requires the production of several virulence factors, including cholera enterotoxin, hemolysin toxin, and HA/protease. | <i>Vibrio cholerae</i> serogroup O1 biotype El Tor (<i>V.cholerae</i> O1 El Tor) was the dominant strain of the 7th global cholera pandemic. It possesses two circular chromosomes that encode for 3,885 total open reading frames (2,770 on Chr 1 and 1,115 on Chr 2). <ref name=S2> [https://doi.org/10.1038/35020000 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.]</ref> Most essential cell function and pathogenicity genes – including those for DNA replication and toxin production – are on Chr 1. <ref name=S2> [https://doi.org/10.1038/35020000 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.]</ref> The ability of V. cholerae to cause illness in hosts requires the production of several virulence factors, including cholera enterotoxin, hemolysin toxin, and HA/protease. | ||
Virulence factors — or pathogenicity factors — are structures, molecules, and regulatory systems that enable pathogens to colonize hosts and counter their immune responses <ref name=S3> [https://doi.org/10.2166/wh.2009.036 Casadevall, Arturo and Pirofski, Liise-anne. "Virulence factors and their mechanisms of action: the view from a damage–response framework." Journal of Water and Health, Vol. 7, Issue S1, 2009, pp. S2–S18.]</ref> | Virulence factors — or pathogenicity factors — are structures, molecules, and regulatory systems that enable pathogens to colonize hosts and counter their immune responses <ref name=S3> [https://doi.org/10.2166/wh.2009.036 Casadevall, Arturo and Pirofski, Liise-anne. "Virulence factors and their mechanisms of action: the view from a damage–response framework." Journal of Water and Health, Vol. 7, Issue S1, 2009, pp. S2–S18.]</ref>. To initiate infection, <i>V.cholerae</i> must traverse the stomach acid and survive among bile and antimicrobial peptides in the small intestines (Ramamurthy et.al, 2020) <ref name=S2> [https://doi.org/10.1038/35020000 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.]</ref> . El Tor strains have as many as 524 virulence-associated genes. Host immune and biological factors induce several of these survival, colonization, and pathogenicity genes <ref name=S2> [https://doi.org/10.1038/35020000 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.]</ref> . | ||
To initiate infection, <i>V.cholerae</i> must traverse the stomach acid and survive among bile and antimicrobial peptides in the small intestines (Ramamurthy et.al, 2020) <ref name=S2> [https://doi.org/10.1038/35020000 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.]</ref> . El Tor strains have as many as 524 virulence-associated genes. Host immune and biological factors induce several of these survival, colonization, and pathogenicity genes <ref name=S2> [https://doi.org/10.1038/35020000 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.]</ref> . | |||
==Cholera Toxin== | ==Cholera Toxin== | ||
Revision as of 02:41, 14 April 2024
Vibrio cholerae O1 biovar El Tor

Vibrio cholerae (V.cholerae) is a gram-negative pathogenic bacterium that infects the small intestine. Symptoms include severe diarrhea, vomiting, and dehydration, which can be life-threatening if not treated promptly. There are more than 200 serogroups of V.cholerae, but only two – O1 and O139 – are associated with cholera epidemics.[1]
V.cholerae is highly motile, with a single polar flagellum that enables it to move quickly in liquid environments. It circulates between the aquatic environment and the human gut. Direct human-to-human transmission of cholera is rare. Instead, the bacteria survive, proliferate, and transmit via contaminated reservoirs, including rivers, ponds, coastal waters, and wells.[1]
Vibrio cholerae serogroup O1 biotype El Tor (V.cholerae O1 El Tor) was the dominant strain of the 7th global cholera pandemic. It possesses two circular chromosomes that encode for 3,885 total open reading frames (2,770 on Chr 1 and 1,115 on Chr 2). [2] Most essential cell function and pathogenicity genes – including those for DNA replication and toxin production – are on Chr 1. [2] The ability of V. cholerae to cause illness in hosts requires the production of several virulence factors, including cholera enterotoxin, hemolysin toxin, and HA/protease.
Virulence factors — or pathogenicity factors — are structures, molecules, and regulatory systems that enable pathogens to colonize hosts and counter their immune responses [3]. To initiate infection, V.cholerae must traverse the stomach acid and survive among bile and antimicrobial peptides in the small intestines (Ramamurthy et.al, 2020) [2] . El Tor strains have as many as 524 virulence-associated genes. Host immune and biological factors induce several of these survival, colonization, and pathogenicity genes [2] .
Cholera Toxin
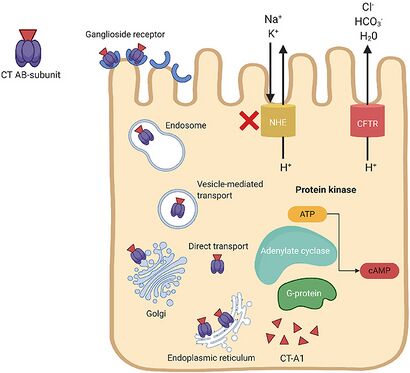
V.cholerae pathogenesis is attributed primarily to the cholera toxin coded for by genes on the core region of the CTXϕ prophage. These genes include ctxA and ctxB, which encode A and B subunits. [1] Cholera toxin (CT) consists of an A subunit (CTA) — responsible for enzymatic activity — and five B subunits (CTB). Once V.cholerae releases the CT complex into the small intestine, the B subunits bind to ganglioside receptors on the surface of intestinal epithelial cells. [1]
Once bound, the CT is endocytosed into the host cell and promptly travels to the Golgi and endoplasmic reticulum. Nicking of a protease-sensitive loop in CTA by host or bacterial protease generates the CT-A1 polypeptide. [4] The catalytic CT-A1 polypeptide leaves the ER and enters the cytosol by retro-translocation, where it activates the Gsα subunit of the guanine nucleotide-binding regulatory (Gαs) protein. [1]
Activation of the Gαs-protein increases adenylate cyclase (AC) activity. AC cleaves ATP to cyclic adenosine monophosphate (cAMP), resulting in uncontrolled increases in intracellular cAMP. Elevated cAMP levels disrupt ion transport mechanisms in the intestinal epithelium. [1]
cAMP activates protein kinase-A (PKA), which inhibits NaCl absorption through the Na+/H+ exchanger and phosphorylates the cystic fibrosis transmembrane conductance regulator (CFTR) chloride channel proteins. [1] This leads to the efflux of chloride ions, HCO3-, Na+, K+, and water into the intestinal lumen. Excessive fluid loss manifests clinically as watery diarrhea, a hallmark symptom of cholera. If left untreated, severe dehydration and electrolyte imbalances can occur, leading to shock, organ failure, and death.
In addition to the cholerae toxin, the accessory cholerae enterotoxin (Ace) and zonula occludens toxin (Zot) contribute to V. cholerae's pathogenesis by inducing changes in the intestinal barrier. Ace appears to cause initial intestinal secretion during infection before the slow-acting CT can. Secretion by Ace utilizes Ca2+ as a second messenger rather than cAMP [5]. However, a comprehensive study on Ace's role in pathophysiology is still lacking.
On the other hand, Zot affects the structure of epithelial tight junctions, which serve as a continuous intercellular barrier between epithelial cells. Zot binds to a cell's α-1-chimaerin receptor and reduces its actin filaments, thus modifying its cytoskeleton and tight junction complex [6]. This modification increases mucosal permeability [7].
Hemolysin Toxin
The ability of V. cholerae to cause illness in hosts requires the production of several virulence factors. For instance, the HlyA gene on Chr 2 codes for a hemolysin toxin. Hemolysin causes blood cell lysis and cytoplasmic vacuolization and is believed to cause lethality and developmental delays in Caenorhabditis elegans infected with V. cholerae (Gao et.al, 2018). HlyA expression is regulated by quorum sensing and regulatory proteins HapR, HlyU, and Fur (Gao et.al, 2018). In the early-to-mid-log phase, HlyU binds to the HlyA promoter to activate its transcription. In late log phase, HapR binds to the Fur, HlyU, and HlyA promoters to repress their transcription (Gao et.al, 2018). HlyA promotes bacterial invasion in the early stages of infection.
HA/protease
V. cholerae also expresses Hemagglutinin/protease (HA/protease). HA/protease is involved in several pathogenic activities, including the modification of other toxins and the degradation of the protective mucus barrier in the gut (Benitez and Silva, 2017). The hapA gene encodes for HA/protease. The transcription of hapA activates under limited nutrient conditions, at high cell population density, and in stationary phase (Benitez and Silva, 2017). Transcription of hapA requires the quorum sensing regulator HapR and RpoS. For example, limited nutrients lead to an increase in intracellular cAMP, which activates RpoS and CRP. CRP then activates HapR (Benitez and Silva, 2017).
toxin-coregulated pilus
Additionally, V. cholerae has become a multidrug-resistant pathogen. The primary cause of V. cholerae drug resistance is the acquisition of extrachromosomal mobile genetic elements (Das, 2020). Sequencing of the genome of clinical and environmental V. cholerae strains has revealed various genetic elements that encode antimicrobial resistance, including integrating conjugative elements, transposable elements, and insertion sequences (Das, 2020).
Conclusion
The genome of Vibrio cholerae enables it to be a highly dangerous pathogen. The bacterium releases various virulence factors, such as hemolysin toxin, cholera toxin, toxin-coregulated pilus, and HA/protease. As the bacterium becomes more multidrug-resistant, Vibrio cholerae will likely continue to pose a worldwide health threat.
Cholera (the disease caused by V. cholerae) has been a public health concern for centuries. Despite sanitation and medical advancements, cholera remains a threat in many areas of the world, particularly those with inadequate sanitation and poor access to clean water. Cholera outbreaks occur regularly in parts of Asia, Africa, and Latin America, and the Middle East. Proper sanitation, clean water, Surveillance systems, and prompt treatment with rehydration therapy are essential for managing and preventing cholera outbreaks.
References
At right is a sample image insertion. It works for any image uploaded anywhere to MicrobeWiki.
The insertion code consists of:
Double brackets: [[
Filename: PHIL_1181_lores.jpg
Thumbnail status: |thumb|
Pixel size: |300px|
Placement on page: |right|
Legend/credit: Magnified 20,000X, this colorized scanning electron micrograph (SEM) depicts a grouping of methicillin resistant Staphylococcus aureus (MRSA) bacteria. Photo credit: CDC. Every image requires a link to the source.
Closed double brackets: ]]
Other examples:
Bold
Italic
Subscript: H2O
Superscript: Fe3+
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Ramamurthy, T. N.,et al. “Virulence Regulation and Innate Host Response in the Pathogenicity of Vibrio cholerae." Frontiers in Cellular and Infection Microbiology, Vol. 10, 2020.
- ↑ 2.0 2.1 2.2 2.3 Heidelberg, J. F.,et al. “DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae.” Nature, Vol. 406, Issue 6795, 2000, pp. 477–483.
- ↑ Casadevall, Arturo and Pirofski, Liise-anne. "Virulence factors and their mechanisms of action: the view from a damage–response framework." Journal of Water and Health, Vol. 7, Issue S1, 2009, pp. S2–S18.
- ↑ Jobling, G. Michael and Holmes, K. Randall. "Identification of motifs in cholera toxin A1 polypeptide that are required for its interaction with human ADP-ribosylation factor 6 in a bacterial two-hybrid system." Proceedings of the National Academy of Sciences of the United States of America, Vol. 97, Issue 26, 2000, pp. 14662–14667.
- ↑ Trucksis, M., et.al. “Vibrio cholerae ACE stimulates Ca(2+)-dependent Cl(-)/HCO(3)(-) secretion in T84 cells in vitro." American Journal of Physiology-Cell Physiology, Vol. 279, No. 3, 2020, pp. C567-C577.
- ↑ Uzzau, S., et al. “Purification and preliminary characterization of the zonula occludens toxin receptor from human (CaCo2) and murine (IEC6) intestinal cell lines." FEMS Microbiology Letters, Vol. 194, Issue 1, 2001, pp. 1–5.
- ↑ Goldblum, E. Simeon., et.al. “The active Zot domain (aa 288–293) increases ZO-1 and myosin 1C serine/threonine phosphorylation, alters interaction between ZO-1 and its binding partners, and induces tight junction disassembly through proteinase activated receptor 2 activation." Federation of American Societies for Experimental Biology, Vol. 25, Issue 1, 2011, pp. 144–158.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski,at Kenyon College,2024
