Bat Influenza A
Introduction
Influenza A is a virus that causes seasonal epidemics almost every winter season in the United States. Influenza A is known to genetically rearrange with influenza viruses from other species to create new strains of the virus, which could have the potential to infect humans. It is important to track the spread of influenza in order to predict future outbreaks and develop vaccines to prevent infection. Influenza A is commonly found in other animals, such as birds and ducks. Influenza A was recently discovered in bats, but it is unknown if bat influenza A virus has the potential to infect humans. However, through viral genetic reassortment, it is possible that bat influenza A can combine genomes with another influenza A virus (such as avian, swine, or human) and cause production of a novel and extremely pathogenic influenza A virus that is able to infect humans very rapidly.
a. Bat ecology and virology
Bats are known to play a crucial role as reservoir hosts of many zoonotic viruses. Zoonotic viruses are viruses that can be passed from an animal to a human. Zoonotic viruses are currently the biggest concern for emerging infections in humans. As of 2012, 75% of emerging infectious diseases was due to zoonoses.
Bats host a large variety of viruses, including lyssaviruses, filoviruses, coronaviruses, and henipaviruses. Bats carry these viruses without being affected by the symptoms, unlike many other mammals. Bats also have far-reaching and overlapping migratory ranges, making intraspecies viral transfer a large factor in their ability to host and spread viruses (Wong et al., 2006).
Researchers all over the world have made great progress in understanding the cause of "spillover events", which are when a virus is transmitted from a bat to a human eventually causing a pandemic. Using models of known bat viruses, researchers have been able to analyze what the drivers of virus emergence are. Halpin et al. (2007) hypothesized that deforestation, roost disturbance, and habitat fragmentation are the main forces that drive bats into urban and periurban areas (Halpin et al., 2007). Continual habitat loss and fragmentation have caused an increased overlap of bat, human, and other animal ecologies. This overlap increases the chances of disease transmission and emergence. Taking these ecological factors into consideration, the potential for bats to host influenza A is of great interest for public health.
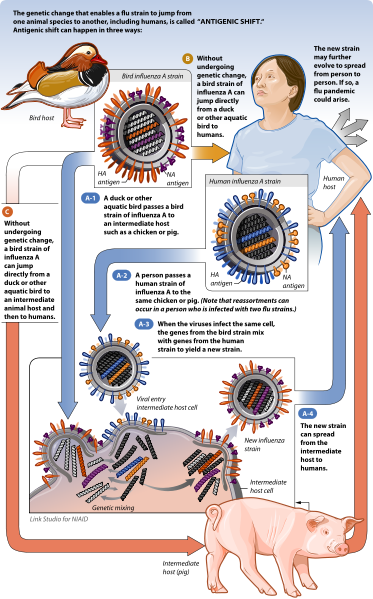
b. Human influenza A
The natural host for influenza A are wild aquatic birds, such as wild ducks, geese, swans, gulls, shorebirds and terns. Influenza A has the potential to infect humans, birds, pigs, horses, dogs, marine mammals, and other animals. The type of influenza is determined by the structure and genetics of two crucial viral proteins- hemagglutinin (HA) and neuraminidase (NA). Currently, there are 17 known HA subtypes and 10 known NA subtypes (Fig. 2). There are various combinations of these proteins that can come together to form different subtypes of influenza viruses (ex. H5N1, otherwise known as avian flu). Many of these subtypes are found to be hosted by animals other than wild aquatic birds, for example, H7N7 is found in horses.
Guatemalan Bat Influenza A (H17N10)
A study done by Tong et al. in 2009-2010 revealed that bats could be a potential reservoir host for influenza A virus. They collected a total of 316 bats from 21 different species across 8 locations in southern Guatemala. They took oral and rectal swab samples from each bat. To test the hypothesis that these bats might harbor influenza viruses, Tong et al. developed a pan-influenza virus RT-PCR that detects the catalytic subunit of RNA polymerase, the polymerase basic protein (PB1). PB1 is one of the most conserved proteins within RNA viruses. By running a PCR reaction with this PB1 RT-PCR primer, they searched for novel influenza viruses within the bat’s genomes. Tong et al. found that three of the 316 bat rectal swab samples, all from the “little-yellow shouldered bat” (Sturnia lilium), were positive for influenza virus. The three samples contained 105-106 viral genome copies per 100 μL of rectal swab suspension, indicating a positive RT-PCR result. Two of the positive samples were captured in 2009 in El Jobo, Guatemala, and the third was captured in 2010 from Agüero, Guatemala (about 50 km away). The viral genomes from the two infected bats GU09-153 and GU09-164 were practically identical to each other (99.99% nucleotide identity). However, they were more distantly related to the infected bat found a year later in Agüero, Guatemala (96.1% nucleotide identity). This shows that the genome of this bat influenza has the potential to change over time and within individual bats.
Tissue samples (liver, intestine, and kidney) taken from the infected bats all tested positive for viral influenza A material, but the oral swab samples tested negative. Tang et al. hypothesizes that this difference in viral material location means that the influenza virus spreads through the bat in an infectious process, compared to orally ingesting infected material. Since the viral material was found within the rectum of the bats, it is most likely that the virus can be contracted through contact with infected bat feces (Tong et al., 2012).
By aligning the nucleotide sequences of known crucial influenza A components, such as hemagglutinin (HA), polymerase (PA), neuraminidase (NA), and nucleoprotein (NP), with the bat influenza viral genome, the identity of the bat virus could be determined. Tong et al. (2012) aligned each of these bat influenza A protein gene segments to known influenza A virus sequences with little to no gaps, with the exception of the NA sequence, which needed 16 gaps to align with influenza A virus (Figure 3). From this data, Tong et al. concluded that the influenza virus discovered in bats in Guatemala is structurally closely related to influenza A.
In order to further classify the subtype of influenza A that these bats host, Tong et al. investigated the genome of the HA protein. By using BLAST, it was found that, on average, the bat virus HA gene has 45% amino acid sequence identity to all HAs from all known influenza A subtypes. In contrast, by looking at the relatedness of NA genes between bat and human influenza A genomes, the bat NA protein only has 24% amino acid sequence identity to other influenza A NA subtypes. The NA protein is involved in the sialic acid-binding/catalytic site, which is how the virus binds to target cells. From this data, Tong et al. concluded that this influenza A virus found is a new subtype, designated as H17. They also concluded that the virus has a different mechanism of infection, and does not use sialic acid-binding sites to bind to target cells.
(Tong et al., 2012).
Peruvian Bat Influenza (H18N11)
Another study done by Tong et al. in 2010 identified another novel influenza A virus, named H18N11, from a flat-faced fruit bat (Artibeus planirostris) in Truenococha, Peru. The intestine tissue from this bat specimen tested strongly positive for influenza virus, while the other tissues tested negative.
A phylogenetic analysis comparing the influenza A genome between the Guatemala and Peru bats showed that the viruses are closely related, but they lie within their own lineages. Excluding the HA gene, the phylogenetic analysis showed that the bat influenza A virus fell outside of the phylogenetic group of all other known influenza A viruses, and that four crucial viral transcription proteins (PB2, PB1, PA and NA) "harbored more genetic diversity than those present in all non-bat (i.e. avian, mammalian) groups combined" (Tong et al., 2013). Considering the outgroup placement of the HA protein gene to the other viral genes, Tong et al (2013) hypothesized that a reassortment event possibly occurred after the viral gene divergence into the bat and non-bat lineages. The HA proteins from one infected bat captured in Peru (A/bat/Peru/10) only had 60.2% sequence identity with the recently identified infected Guatemalan H17 influenza A virus. This data supports the designation of the Peruvian bat influenza A to be a novel H18 subtype.
Potential for Human Infection
The detection of this virus in 1% of the total bat population tested in Guatemala correlates to the frequency of influenza virus detection in wild bird populations, which are known to commonly transmit influenza to humans (Munster et al., 2007). This means that influenza A is as prevalent in those bat colonies as it is in certain wild bird populations. However, by investigating the crucial structural components of influenza A and comparing their genes to the bat influenza virus, it was concluded by Tang et al. (2013) that the influenza virus from both Guatemalan and Peruvian bats are not able to infect humans directly. This hypothesis was tested by using a mini-genome reporter plasmid, which contains a luciferase gene that is driven by non-coding regions from bat (Guat/164.NS-NCR) or human (WSN.NS-NCR) influenza virus. This vector showed high levels of viral transcription when co-transfected with a complete polymerase gene set from bat influenza virus. The viral transcription of H17 (Guatemala) was 5 fold higher than H18 (Peru). In contrast, H18 bat influenza was 16-fold less effective at virus transcription when co-transfected with the human influenza virus vector, indicating that the bat influenza might not be able to effectively interact with the human influenza virus, making genetic reassortment unlikely. Attempts to propagate bat influenza in mammalian and avian cell cultures were also unsuccessful (Tong et al., 2013).
a.Viral genetic reassortment
RNA viruses are extremely diverse due to their high mutation rates. The error-prone nature of RNA synthesis causes many RNA viruses to mutate very easily. Influenza viruses, which have segmented genomes, are diverse because of genetic reassortment. During influenza infection, individual RNA segments enter the cell's nucleus to be copied numerous times to form new RNA genomes for new virions. The new RNA segments are then transported to the cytoplasm and then are packaged inside the new virions, which then bud off from the cell to infect other cells. This process becomes interesting when a cell is infected with two different times of influenza viruses, where both of the RNA genomes from both viruses are being copied at the same time in the host cell's nucleus. When the new virions are being assembled, it is very possible that each of the 8 RNA segments could come from either virus. The virions that contain RNA segments from two different virus strains are called "reassortments". A classic example of viral reassortment was when influenza genomes reassorted to form H1N1, which caused a global pandemic in 2009 (Figure 5). H1N1 is a viral reassortment of avian, human, and swine influenza viruses. However, reassortment can only occur between influenza viruses of the same type, i.e. influenza A with influenza A. It is not known why influenza A viruses do not exchange RNA segments with influenza B or C strains, and vice versa (Trifonov, et. al., 2009).
One Health Approach
a. Programs for the detection, identification, and prevention of bat virus pandemics
A "One Health" program implements a collaborative effort from multiple disciplines, such as public health officials, veterinarians, and conservation biologists, to effectively prevent pandemic events. One Health initiatives have been shown to be effective for many zoonotic virus cases. Organizations like the World Health Organization (WHO), the Center for Disease Control (CDC) and EcoHealth Alliance are extremely useful in preventing pandemics. For example, the World Health Organization initiated response teams after the recognition of SARS to inform the public and form quarantines in infected areas (Smith et al., 2012).
There are many One Health programs in place for the prevention of emerging infectious diseases, including bat zoonotic viruses. The Sicki Project, organized by EcoHealth Alliance, is aimed to “unravel the origins of Emerging Infectious Diseases and outbreaks by curating, reviewing, and expanding the scope of the data used by the global health community to model disease emergence” ([6], The Sicki Project). The project has helped to estimate areas in the world where there is high risk of infectious disease emergence (Figure 10). EcoHealth Alliance is at the front line of viral discovery, already having aided in discovering over 100 novel bat viruses around the world.
b. Bat conservation
Bats are diverse and interesting mammals, and they are also crucial components of ecosystems. For example, they are important pollinators for many ecosystems all over the world. Unfortunately, many bat species have become locally extinct because of habitat loss, cave disturbances, proliferation of wind-energy facilities, bush meat overhunting, and pesticide use (Kunz et. al., 2011) .Bats are unique and beneficial animals, yet they are the one of the least studied and most misunderstood of animals.
Bats are increasingly becoming recognized as reservoir hosts for many zoonotic viruses. The rate of emergence of zoonotic viruses appears to be increasing, and we are becoming more efficient at investigating the reasons why these viruses emerge. Ecological drivers, such as land use change, agricultural intensification, poor public health practices, and increasing urbanization, among others, are reasons why these viruses can jump from a reservoir host to humans.
Further Work
There is much work to be done researching bat influenza A. First, it is crucial that we understand how the virus binds to cell receptors. It has been hypothesized by Sun et. al. (2013) that since the bat-derived H17 protein does not bind to canonical avian or human receptors, it must use a different entry mechanism. They discovered the crystalline structure of H17 and found that it has special features, including a distorted sialic acid (SA) binding site and low thermostability (Sun et al., 2013 [7]).
Another study was recently published in 2013 reporting data on the structure of the PA protein of H17N10. Tefsen et. al. found that the PA protein in bat influenza A has similar fold structure as homologous PA domains found in known influenza A strains. They also found that it demonstrates endonuclease activity and a histidine residue located in the active site is essential for that activity. They conclude that even though the bat influenza A is not infectious to humans, it is very possible that the virus could genetically reassort with canonical influenza viruses (Tefsen et. al., 2013)[8].
References
1. Halpin, K., Hyatt, A. D. Emerging viruses: Coming in on a Wrinkled Wing and a Prayer. 2007. Emerging Infections CID: 44: 711-717. [[9]]
2. Kunz, T. H., Braun de Torrez, E., Bauer, D., Lobova, T., Fleming, T. H. Ecosystem services provided by bats. 2011. Ann. N.Y. Acad. Sci.: 1223, 1-38. [[10]]
3. Munster, V. J., Bass, C., Lexmond, P., Waldenström, J., Wallensten, A., Fransson. T., Rimmelzwaan, G. F., Beyer, W. E., Schutten, M., Olsen, B., Osterhaus, A. D., Fouchier, R. . Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. 2007. PLoS Pathog 3(5): 630-638.[[11]]
4. Sun, X., Shi, Y., Lu, X., He, J., Gao, F., Yan, J., Qi, J., Gao, G. F. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. 2013. Cell Rep. 3(3): 769-778. [[12]]
5. Tefsen, B., Lu, G., Zhu, Y., Haywood, J., Zhao, L., Deng, T., Qi, J., Gao, G. The N-terminal domain of PA from bat-derived influenza-like virus H17N10 has endonuclease activity. 2013. Journal of Virol. Published ahead of print doi: 10.1128/JVI.03270-13 [13]
6. Tong, S., Li, P., Rivailler, C., Conrardy, C., Alvarez-Castille, D., Chen, L., Recuenco, S., Ellison, J. A., Davis, C. T., York, I. A., Turmelle, A. S., Moran, D., Rogers, S., Shi, M., Tao, Y., Weil, M. R., Tang, K., Rowe, L. A., Sammons, S., Xiyan, X., Frace, M., Lindblade, K. A., Cox, N. J., Anderson, L. J., Rupprect, C. E., and Donis, R. O. A distinct lineage of influenza A virus from bats. 2012. PNAS, 109 (11): 4269-4274. [[14]]
7. Tong S., Zhu, X., Mang, S., Zhang, J., Bourgeois, M., Yang, H., Chen, X., Rucuenco, S., Gomez, J., Chen, L., Johnson, A., Tao, Y., Dreyfus, C., Yu, W., McBride, R., Carney, P., Gilbert, A., Chang, J., Guo, Z., Davis, C., Paulson, J. C., Stevens, J., Rupprecht, C. E., Holmes, E. C., Wilson, I. A., Donis, R. O. New World Bats Harbor Diverse Influenza A Viruses. PLOS: Pathogens. 9(10): e1003657 [[15]]
8. Trifonov, V., Khiabanian, H., Rabadan, R. Geographic Dependence, Surveillance, and Origins of the 2009 Influenza A (H1N1) Virus. 2009. N. Engl. J. Med.: (361): 115-119 [16]
9. Wong, S., Lau, S., Woo, P., Yuen, K. Y. Bats as a continuing source of emerging infections in humans. 2006. Rev. Med. Virol. 17: 67-91. [[17]]