Gastritis and Peptic Ulcer Disease Caused by Helicobacter pylori
Introduction
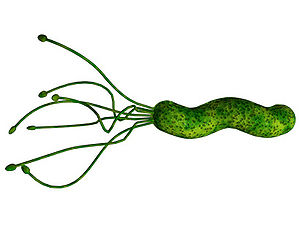
Helicobacter pylori Figure 1. is a Gram-negative, helix-shaped bacterium that is about 3 micrometers long with a diameter of 0.5 micrometers. H. pylori is a microaerophilic bacterium which means that it requires oxygen to function. However, H. pylori requires much lower concentrations of oxygen than those found in our atmosphere. This bacterium contains a hydrogenase which it can use to obtain energy by oxidizing molecular hydrogen (in the form of H2) produced by intestinal bacteria. H. pylori also produces oxidase, catalase, and urease. It has an outer-membrane consisting of phospholipids and lipopolysaccharide which are characteristic of typical Gram-negative bacteria. H. pylori bacteria are known to inhabit various areas of the stomach and duodenum. Infections caused by this bacteria lead to chronic inflammation in the stomach lining (Gastritis). H. pylori infections are also strongly associated to the development of gastric ulcers and even stomach cancer. Although H. pylori is known to cause several problems, over 80% of individuals infected with this bacterium show no symptoms. Over half of all people in the world are thought to harbor this bacterium in their upper gastrointestinal tract.[14]
Pathogenesis
There is no single pathway of transmission for Helicobacter pylori that has been clearly identified. Scientists have had difficulty locating the bacterium anywhere outside of gastric tissue. Because of this, it has been difficult for scientists to locate the portal by which the bacterium is spread. However, scientists do believe the bacterium is passed from person to person and most transmissions occur during early life. It has been suggested that transmission may occur in faecal-oral and oral-oral pathways. Other pathways have not been ruled out.[25] However the bacterium enters the body it eventually makes its way to the gastrointestinal tract. H. pylori makes its way into the stomach to begin colonization. In order to colonize the stomach H. pylori must survive the acidic pH of its environment. It burrows into the mucous lining that coats the stomach. An H. pylori bacterium has flagella and uses them to move through the stomach lumen and drill into the mucous lining of the stomach.[1] H. pylori must insert itself near the stomach's epithelial cell layer Figure 2. which is this bacterium’s niche. H. pylori can be found deep in the mucous lining of the stomach. Mucus is continuously secreted by mucous cells and removed on the side of the lumen. To avoid being carried into the lumen by the newly formed mucus, H. pylori senses the pH gradient within the mucus layer by chemotaxis and swims away from the acidic contents of the lumen towards the more neutral pH environment of the epithelial cell surface.[2] H. pylori can also be present on the inner surface of the epithelial cells of the stomach. Sometimes the bacterium is even found inside the epithelial cells.[3] H. pylori produces adhesins that bind to membrane-associated lipids and carbohydrates which helps it to adhere to the epithelial cells.[4] The bacterium also produces great quantities of urease, and enzyme located inside and outside of the cell. This enzyme is capable of breaking down urea that is secreted into the stomach. It breaks down the urea into carbon dioxide and ammonia. The ammonia is converted into an ammonium ion by accepting a hydrogen ion from self-ionized water. The left-over hydroxyl ions then react with the carbon dioxide to produce bicarbonate which neutralizes gastric acid. Therefore, H. pylori is dependent upon urease for its survival in the acidic environment found in the stomach. Without the urease enzyme the bacterium would almost inevitably die. The ammonia that is a byproduct of the urease reaction is toxic to epithelial cells of the stomach. H. pylori also produces protease (an enzyme that breaks down proteins), vacuolating cytotoxin A (VacA), and phospholipases (enzymes that hydrolyze phospholipids into fatty acids and other lipophilic substances). All of these products are damaging to the epithelial cells of the stomach.[5]
When H. pylori colonizes the stomach it often results in chronic gastritis, an inflammation of the stomach lining. Stomach and duodenal ulcers occur when the inflammation allows the acid and pepsin in the stomach lumen to overpower the mechanisms that protect the stomach and duodenal mucosa from these substances. The location of the chronic gastritis (which occurs at the site of H. pylori colonization) dictates the type of ulcer that subsequently develops.[6] The amount of acid within the stomach lumen has an effect on the colonization patterns of the bacterium H. pylori. It will therefore, in the end, establish the location at which the gastric or duodenal ulcer will form. For example, in people that produce large amounts of acid, H. pylori will colonize the pyloric antrum of the stomach to avoid the parietal cells in the corpus of the stomach that secrete acid.[7]
The inflammatory response to the bacteria induces G cells in the antrum to secrete the hormone gastrin. Gastrin travels through the bloodstream to the corpus of the stomach.[8] Gastrin excites the parietal cells in the corpus to release even more acid into the stomach lumen. Persistently increased gastrin levels eventually cause the number of parietal cells to also increase, further increasing the amount of acid secreted.[9] The increased amount of acid damages the duodenum and may consequently result in the formation of duodenal ulcers. Gastric ulcers on the other hand are often linked with reduced or normal levels of acid production. This suggests that the mechanisms that protect the gastric mucosa are faulty in that individual.[9] H. pylori capitalizes on this defect and begins to colonize the corpus of the stomach where the parietal cells are located. The chronic inflammation caused by the colonization of this bacterium causes further reduction in the stomach’s acid production, causing atrophy of the stomach lining. The deterioration of the stomach’s lining may lead to future gastric ulceration and even an increased risk for stomach cancer.[10]
There are some strains of H. pylori that are capable of triggering a greater inflammatory response in the stomach of its host. This strain carries the cag pathogenicity island (cag PAI). Over half of the H. pylori strains in Western countries are thought to carry the cag PAI. [11] Not only do these strains create a stronger inflammatory response in the stomach but they also create a greater risk for developing ulcers or cancer than the strains lacking the cag PAI.[7] The cag PAI expresses a type IV secretion system after the attachment of the bacterium to the epithelial cells of the stomach. This system inserts peptidoglycan from the bacterial cell wall into the epithelial cells. This peptidoglycan acts as an inflammatory inducing agent within epithelial cells. It is recognized by the cytoplasmic immune sensor Nod1 that stimulates the expression of cytokines which promote inflammation.[12]
The type IV secretion system also injects the cag PAI-encoded protein CagA into the stomach's epithelial cells, where it disrupts the cytoskeleton, adherence to adjacent cells, intracellular signaling, cell polarity and other cellular activities.[13] Once it reaches the inside of the cell the CagA protein is phosphorylated on tyrosine residues by a host cell membrane-associated tyrosine kinase. Pathogenic strains of H. pylori have been shown to activate the epidermal growth factor receptor (EGFR), a membrane protein with a tyrosine kinase domain. Activation of the EGFR by H. pylori is linked with altered signal transduction and gene expression in host epithelial cells that may contribute to pathogenesis. It has also been suggested that a c-terminal region of the CagA protein (amino acids 873–1002) is able to regulate host cell gene transcription independent of protein tyrosine phosphorylation.[14][15] There is a large amount of diversity between different strains of H. pylori, and often times the strain with which a person is infected is predictive of the outcome of the infection.
There are two possible mechanisms currently being investigated by which H. pylori could cause cancer. One of these mechanisms involves the enhanced production of free radicals near H. pylori as well as an increased rate of host cell mutation.The other mechanism under investigation is called the "perigenetic pathway"[16] and involves enhancement of the transformed host cell phenotype by altering cell proteins such as adhesion proteins. It is thought that H. pylori causes inflammation and locally high levels of TNF-α and/or interleukin 6. According to the perigenetic mechanism, inflammation-associated signaling molecules such as TNF-α can alter gastric epithelial cell adhesion and cause the dispersal and relocation of mutated epithelial cells without the need for additional mutations in tumor suppressor genes such as genes that code for cell adhesion proteins.[17]
Research
From the mid 1990s to around the year 2000 a group of scientists conducted an experiment to investigate variation within the cag pathogenicity island (PAI) of Helicobacter pylori and to evaluate the effect on expression of anti-CagA antibody. The bacteria were isolated from patients with dyspepsia in mid-Essex.[31] During the experiment sixty two isolates of H. pylori were cultured from gastric biopsies and were screened by specific PCR assays for the presence of cagA and other gene markers (cagD and cagE, and virD4) in the cag PAI. An enzyme linked immunosorbent assay (ELISA) kit (Viva Diagnostica helicobacter p120) was used to test for anti-CagA IgG antibody in matching sera. The isolates were also genotyped by vacuolating cytotoxin polymerase chain reaction (PCR) analysis, and tested for absence of the complete cag PAI (empty site PCR assay). The experiment resulted with forty one of the H. pylori isolates having a cag PAI containing cagA. One strain had no cagA but other cag PAI loci were present, whereas the remaining 20 strains had no detectable cag PAI markers. Anti-CagA IgG antibody was detected in 34 sera by the ELISA assay, and when compared with the cag PAI genotype of the infecting strain, accuracy, sensitivity, and specificity were 92%, 87%, and 100%, respectively. The seven discrepant or borderline strains in the ELISA were all vacA s1 but differed in other genotypic markers.[31] The scientists concluded that the cag PAI was widely distributed in H. pylori from patients with dyspepsia in mid-Essex who had different gastric pathologies. Infection with a strain having an uninterrupted cag PAI was associated with the presence of anti-CagA antibody in most patients. Discrepant ELISA results, mostly for elderly patients with duodenal ulcers, were attributed to cagA associated variation, particularly to the presence of mixed cagA+/cagA− cell variants in the infecting strain population. Tests for anti-CagA serum antibody were unreliable for predicting severity of clinical disease associated with H. pylori infection in this series of patients.[31]
Diagnosis
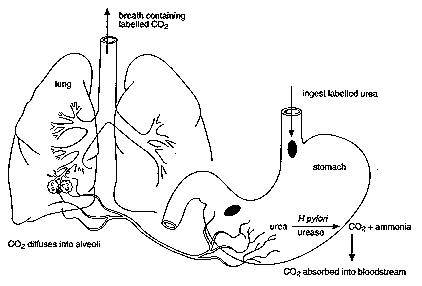
Diagnosis often begins by checking the patient for symptoms of dyspepsia (upset stomach/indigestion). This problem can often indicate an H. pylori infection. After dyspeptic symptoms are noticed there are more specific tests that can be used to identify whether a person has an H. pylori infection or not. Many of these tests are non-invasive. Three of these non-invasive tests include; a blood antibody test, a stool antigen test, or a carbon urea breath test. Figure 4. There are different advantages and limitations for each of these tests. For example, blood antigen tests have a 76%-84% sensitivity. However, they are limited due to the fact that they do not test for the bacterium itself but rather for the antibody your body creates as an immune response. Therefore, a false positive could occur if one had been infected by H. pylori in the past. They might not currently have the bacterial infection but because that person had the infection at a previous time there would still be antibodies present in their blood stream. In a carbon urea breath test the patient drinks urea which contains labeled carbons (13C,14C) which is metabolized by the bacterium (using urease enzyme) producing labeled carbon dioxide that can be detected in the patient’s breath.[18] This test can sometimes give false negatives because certain drugs can affect the ability of H. pylori‘s urease to function properly.[19] Another effective test is the urine ELISA test with a 96% sensitivity and 79% specificity. The most reliable test for identifying H. pylori infection involves a biopsy check during endoscopy with a rapid urease test, a histological examination, and a microbial culture. None of the testing methods are completely failsafe.
Treatment
After H. pylori is identified in patients with gastritis or a peptic ulcer, the standard procedure is to eradicate the bacterial infection and allow the ulcer to heal. The typical therapy is a one week “triple therapy” consisting of a proton pump inhibitor such as omeprazole Figure 5. and the antibiotics clarithromycin and amoxicillin.[20] There have been different varieties of the “triple therapy” that have been developed. Some varieties may use a different proton pump inhibitor such as pantoprazole or rabeprazole. They may also replace the amoxicillin with metronidazole for people who are allergic to penicillin.[21] The new revolutions in therapy have made the treatment of peptic ulcers easier and have made it possible to cure the disease. In prior times only the symptoms were treated using antacids, H2-antagonists or proton pump inhibitors alone. [22]Now further advancements have allowed for the treatment process to even decrease from 14 days to a possible 7-10 day treatment period. With advances in the efficacy of proton pump inhibitors a change in the patients diet is not even necessary. An article in The American Journal of Clinical Nutrition gives evidence that probiotics such as lactic acid bacteria can actual inhibit H. pylori infections. The article states that "ingesting lactic acid bacteria exerts a suppressive effect on H. pylori infection in both animals and humans," stating that "supplementing with Lactobacillus- and Bifidobacterium-containing yogurt (AB-yogurt) was shown to improve the rates of eradication of H. pylori in humans."[23] The fact that more and more people infected with H. pylori are found to harbor antibiotic-resistant bacteria poses a problem for treatment of the infection. This scenario inevitably results in initial treatment failure and requires doctors to attempt using additional rounds of antibiotic therapy or alternative strategies. One such strategy is called “quadruple therapy”, which adds a bismuth colloid, such as Bismuth subsalicylate.[18] For the treatment of clarithromycin-resistant strains of H. pylori, levofloxacin use is suggested.[24]
Prevention

Increasing amounts of antimicrobial resistance in H. pylori have created the need for a prevention strategy. Scientists have conducted extensive vaccine studies using mouse models that have shown promising results.[26] Researchers are studying different methods of immunization (such as using antigens) to find the best system of immune protection. Most of the research has only recently been moving from animal to human trials.[27]An intramuscular vaccine against infection is undergoing Phase I clinical trials and has shown an antibody response against H. pylori. Its clinical usefulness requires further study.[28] A 2009 study found that green tea may be able to prevent inflammation if it is ingested prior to exposure to Helicobacter infection. Recent studies have also shown that H. pylori activity can be inhibited through other dietary measures. In 2009 a Japanese study found that eating as little as 70 g (2.5 ounces) of broccoli sprouts daily for two months reduces the number of colonies of H. pylori bacteria in the stomach by 40% in mice and humans. The treatment also seemed to help by enhancing the protection of the gastric mucosa against H. pylori. However, it was relatively ineffective on related gastric cancers. The previous bacterial infection returned within two months after broccoli sprouts were removed from the diet. Therefore, an ongoing diet of broccoli sprouts is best for continued protection from H. pylori.[30] Another study published in 2008 in the Korean Journal of Microbiology and Biotechnology found that kimchi (a traditional Korean dish) Figure 6. contains a strain of bacteria "showing strong antagonistic activity against H. pylori." The bacterium strain isolated from kimchi, designated Lb. plantarum NO1, was found to reduce the urease activity of H. pylori by 40-60% and suppress H. pylori’s binding to human gastric cancer cell line by more than 33%.[29]
Conclusion
There is a plethora of literature concerning gastritis and peptic ulcer disease caused by the bacterium Helicobacter pylori. Nevertheless, there is still much to be learned about this bacterium and its effects on the human body. It may not be known exactly how H. pylori is transmitted but at least we are able to detect and eradicate the bacterium with relative ease and efficiency. Many new ways to help prevent and inhibit the activity of H. pylori are being discovered. Now it is up to the scientists to discover even better ways to treat the disease caused by this bacterium and to find ways to prevent the disease. When H. pylori’s mode of transmission is finally discovered, it may lead to more efficient ways to prevent transmission and infection.
References
1. Ottemann KM, Lowenthal AC (April 2002). "Helicobacter pylori uses motility for initial colonization and to attain robust infection". Infect. Immun. 70 (4): 1984–90. doi:10.1128/IAI.70.4.1984-1990.2002. PMID 11895962. PMC 127824. http://iai.asm.org/cgi/pmidlookup?view=long&pmid=11895962.
2. Schreiber S, Konradt M, Groll C, et al. (April 2004). "The spatial orientation of Helicobacter pylori in the gastric mucus". Proc. Natl. Acad. Sci. U.S.A. 101 (14): 5024–9. doi:10.1073/pnas.0308386101. PMID 15044704.
3. Petersen AM, Krogfelt KA (May 2003). "Helicobacter pylori: an invading microorganism? A review". FEMS Immunol. Med. Microbiol. 36 (3): 117–26. doi:10.1016/S0928-8244(03)00020-8. PMID 12738380.
4. Ilver D, Arnqvist A, Ogren J, et al. (January 1998). "Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging". Science (journal) 279 (5349): 373–7. PMID 9430586.
5. Smoot DT (December 1997). "How does Helicobacter pylori cause mucosal damage? Direct mechanisms". Gastroenterology 113 (6 Suppl): S31–4; discussion S50. PMID 9394757.
6. Dixon MF (February 2000). "Patterns of inflammation linked to ulcer disease". Baillieres Best Pract Res Clin Gastroenterol 14 (1): 27–40. doi:10.1053/bega.1999.0057. PMID 10749087.
7. Kusters JG, van Vliet AH, Kuipers EJ (July 2006). "Pathogenesis of Helicobacter pylori infection". Clin Microbiol Rev 19 (3): 449–90. doi:10.1128/CMR.00054-05. PMID 16847081.
8. Blaser MJ, Atherton JC (February 2004). "Helicobacter pylori persistence: biology and disease". J. Clin. Invest. 113 (3): 321–33. doi:10.1172/JCI20925. PMID 14755326.
9. Schubert ML, Peura DA (June 2008). "Control of gastric acid secretion in health and disease". Gastroenterology 134 (7): 1842–60. doi:10.1053/j.gastro.2008.05.021. PMID 18474247.
10. Suerbaum S, Michetti P (October 2002). "Helicobacter pylori infection". N. Engl. J. Med. 347 (15): 1175–86. doi:10.1056/NEJMra020542. PMID 12374879.
11. Peek RM, Crabtree JE (January 2006). "Helicobacter infection and gastric neoplasia". J. Pathol. 208 (2): 233–48. doi:10.1002/path.1868. PMID 16362989.
12. Viala J, Chaput C, Boneca IG, et al. (November 2004). "Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island". Nat. Immunol. 5 (11): 1166–74. doi:10.1038/ni1131. PMID 15489856.
13. Backert S, Selbach M (August 2008). "Role of type IV secretion in Helicobacter pylori pathogenesis". Cell. Microbiol. 10 (8): 1573–81. doi:10.1111/j.1462-5822.2008.01156.x. PMID 18410539
14. Baldwin DN, Shepherd B, Kraemer P, et al. (February 2007). "Identification of Helicobacter pylori genes that contribute to stomach colonization". Infect Immun 75 (2): 1005–16. doi:10.1128/IAI.01176-06. PMID 17101654.
15. Broutet N, Marais A, Lamouliatte H, et al. (April 2001). "cagA Status and eradication treatment outcome of anti-Helicobacter pylori triple therapies in patients with nonulcer dyspepsia". J Clin Microbiol 39 (4): 1319–22. doi:10.1128/JCM.39.4.1319-1322.2001. PMID 11283049. PMC 87932. http://jcm.asm.org/cgi/pmidlookup?view=long&pmid=11283049.
16. Tsuji S, Kawai N, Tsujii M, Kawano S, Hori M (July 2003). "Review article: inflammation-related promotion of gastrointestinal carcinogenesis--a perigenetic pathway". Aliment. Pharmacol. Ther. 18 Suppl 1: 82–9. doi:10.1046/j.1365-2036.18.s1.22.x. PMID 12925144.
17. Suganuma M, Yamaguchi K, Ono Y, et al. (July 2008). "TNF-α-inducing protein, a carcinogenic factor secreted from H. pylori, enters gastric cancer cells". Int. J. Cancer 123 (1): 117–22. doi:10.1002/ijc.23484. PMID 18412243.
18. Stenström B, Mendis A, Marshall B (August 2008). "Helicobacter pylori - The latest in diagnosis and treatment". Aust Fam Physician 37 (8): 608–12. PMID 18704207
19. Logan RP, Walker MM (October 2001). "ABC of the upper gastrointestinal tract: Epidemiology and diagnosis of Helicobacter pylori infection". BMJ 323 (7318): 920–2. doi:10.1136/bmj.323.7318.920. PMID 11668141.
20. Mirbagheri SA, Hasibi M, Abouzari M, Rashidi A (August 2006). "Triple, standard quadruple and ampicillin-sulbactam-based quadruple therapies for H. pylori eradication: a comparative three-armed randomized clinical trial". World J. Gastroenterol. 12 (30): 4888–91. PMID 16937475. http://www.wjgnet.com/1007-9327/12/4888.asp. Retrieved 2008-09-02.
21. Malfertheiner P, Megraud F, O'Morain C, et al. (June 2007). "Current concepts in the management of Helicobacter pylori infection: the Maastricht III Consensus Report". Gut 56 (6): 772–81. doi:10.1136/gut.2006.101634. PMID 17170018.
22. Graham DY, Lew GM, Evans DG, Evans DJ, Klein PD (August 1991). "Effect of triple therapy (antibiotics plus bismuth) on duodenal ulcer healing. A randomized controlled trial". Ann. Intern. Med. 115 (4): 266–9. PMID 1854110.
23. Kuan-Yuan Wang , “Effects of ingesting Lactobacillus- and Bifidobacterium-containing yogurt in subjects with colonized Helicobacter pylori1,2,3” American Journal of Clinical Nutrition
24. Perna F, Zullo A, Ricci C, Hassan C, Morini S, Vaira D (November 2007). "Levofloxacin-based triple therapy for Helicobacter pylori re-treatment: role of bacterial resistance". Dig Liver Dis 39 (11): 1001–5. doi:10.1016/j.dld.2007.06.016. PMID 17889627.
25. Goodman K, Correa P (March 1995). “The Transmission of Helicobacter pylori. A Critical Review of the Evidence” International Journal of Epidemiology.
26. Hoffelner H, Rieder G, Haas R (January 2008). "Helicobacter pylori vaccine development: optimisation of strategies and importance of challenging strain and animal model". Int. J. Med. Microbiol. 298 (1–2): 151–9. doi:10.1016/j.ijmm.2007.07.006. PMID 17714988.
27. Kabir S (April 2007). "The current status of Helicobacter pylori vaccines: a review". Helicobacter 12 (2): 89–102. doi:10.1111/j.1523-5378.2007.00478.x. PMID 17309745.
28. Malfertheiner P, Schultze V, Rosenkranz B, et al. (May 2008). "Safety and Immunogenicity of an Intramuscular Helicobacter pylori Vaccine in Noninfected Volunteers: A Phase I Study". Gastroenterology 135 (3): 787. doi:10.1053/j.gastro.2008.05.054. PMID 18619971.
29. Youl Lee and Hae Choon Chang (2008). "Isolation and Characterization of Kimchi Lactic Acid Bacteria Showing Anti-Helicobacter pylori Activity". Korean Journal of Microbiology and Biotechnology 2: 106–114. http://kmbase.medric.or.kr/Main.aspx?d=KMBASE&m=VIEW&i=1234420080360020106.
30. Yanaka et al.; Fahey, JW; Fukumoto, A; Nakayama, M; Inoue, S; Zhang, S; Tauchi, M; Suzuki, H et al. (April 2009). "Dietary Sulforaphane-Rich Broccoli Sprouts Reduce Colonization and Attenuate Gastritis in Helicobacter pylori-Infected Mice and Humans". Cancer Prevention Research 2 (4): 353–360. doi:10.1158/1940-6207.CAPR-08-0192. PMID 19349290. http://cancerpreventionresearch.aacrjournals.org/cgi/content/abstract/2/4/353
31. Peters TM, Owen RJ, Slater E, Varea R, Teare EL, Saverymuttu S (August 2000) “Genetic diversity in the Helicobacter pylori cag pathogenicity island and effect on expression of anti-CagA serum antibody in UK patients with dyspepsia” Journal of Clinical Pathology.
