Planococcus halocryophilus; growth in subzero halophilic conditions
Planococcus halocryophilus (OR1)
By Ethan Hanson
Classification:
Kingdom: Bacteria
Phylum: Firmicutes
Class: Bacilli
Order: Bacillaes
Family:Planococcaceae
Genus: Planococcus
Species:
Planococcus halocryophilus
Planococcus halocryophilus is an aerobic, gram-positive bacterium that is found in arctic permafrost. This extremophile is characterized as both halophilic and psychrophilic, thriving in an environment of high salinity as well as an extremely low temperature. This bacterium's reproduction capability is measured at the lowest recorded temperature, measured at -15ºC. Planococcus halocryophilus continues to preserve itself at temperatures as low as -25ºC. The bacterium is accountable for effects of global warming, bringing about sizable CO2 emissions concurrent to melting permafrost. Astrobiological research towards this extremophile is resonant owing to its similitude of potential target environments for life on Mars.
[1]
Cell Structure and Metabolism
Planococcus halocryophilus (OR1) cells are non-spore forming motile cocci that occur individually or in pairs, with a diameter of 0.8-1.2 micrometers. In colonies, P. halocryophilus appears an opaque, shiny orange color in clusters measured between 2.0-3.0 mm. As stated before, they are gram-positive, aerobic, and both psychrotolerant as well as halotolerant. They can grow at temperatures between -15 and 37ºC, however their optimal temperature of growth is approximately 25ºC.
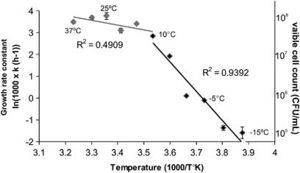
Figure 1 exhibits an Arrhenius plot of log transformed growth rate constants for OR1. Individual measurements come from viable cell count in relation to temperature. Here, the optimal temperature of growth is projected at 25ºC with a maximum 10^8 colony forming units per mL, and a significant drop in growth rate when temperature falls below 10ºC. At <10ºC, the R2 value (slope) increases from approximately 0.49 to approximately 0.94. Growth rate seems to negate below -10ºC. [2][3]
P. halocryophilus are capable of growth in liquid water of high salinity, up to 19% NaCl, but maintain the ability to grow in conditions of 0% NaCl. When compared to other strains of Planococcus, P. halocryophilus was the only species with a growth temperature range below zero. OR1 also measured out to be the most tolerant to NaCl (ahead of P. maritimus at 17% NaCl). A major physiological feature that distinguishes P. halocryophilus is its transformation of the cellular envelope at temperatures below zero. OR1 cells grown at subzero temperature gain winding nodular sheet encrustations around each individual cell. Its hypothesized that the purpose of this cellular envelope addition is to provide extra protection to the interior of the cell during low-temperature replication.
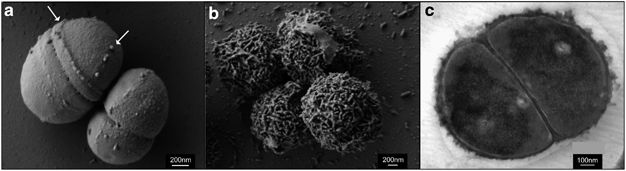
Figure 2 shows three electron microscopic images of P. halocryophilus, (a) and (b) under scanning EM and (c) under transmission EM. Images (a) and (c) show comparison of cell division planes, for cells replicating at 25ºC (a), versus dividing cells at -15ºC (c). The images show dramatic contrast by the nodular cell envelope, showing a 3-dimensional representation of the encrustation in image (c). [4][5]
P. halocryophilus is an aerobic heterotroph that has the ability to utilize at least 25 different carbon sources (glucose, fructose, galactose, glycerol, rhamnose, mannose, glucosamine, glutamine, maltose, ribose, cellobiose, trehalose, mannitol, lactose, lactic acid, sucrose, gelatin, pectin, dextrin, serine, alanine, arginine, acetic acid, gluconic acid, and glutamic acid), when provided individually. [6]
A cellular fatty acid profile of P. halocryophilus was used to determine which fatty acids were most abundant under specific conditions (control at 25ºC and 0% NaCl) of high salinity and of subzero temperatures separately. Under conditions of high salinity, the most abundant were branched fatty acids and unsaturated fatty acids. Under subzero temperature conditions, the most abundant were straight-chain fatty acids and saturated fatty acids. Very few outliers showed greatest abundance in control conditions. This data is important to membrane fluidity of P. halocryophilus because of the relative abundance of saturated and unsaturated fatty acids. Having more unsaturated fatty acids in conditions of high salinity provides evidence for higher fluidity in the cell membrane, allowing for greater osmotic regulation in ideal temperature conditions. In contrast, the high abundance of saturated fatty acids in subzero conditions provide evidence for lower cell fluidity, in order to protect the cell when metabolic energetics are low due to temperature. [7][8]
In conditions of cold-specific stress, energy metabolism carries on through increased abundance of substrate shuttling genes and components of the electron transport chain. In subzero conditions (-15ºC), while most metabolic enzymes are repressed, the tricarboxylic cycle and ATP synthase keep the cell alive. In conditions of salt-specific stress, carbohydrate metabolism is extricated by glycogen synthesis, stores induced by specific genes that are activated only under nutrient limited conditions. P. halocryophilus proves its unique resourcefulness from specific response methods through fastidious induction and regulation of metabolic activity. [9]
Genome Structure
Include some current research, with at least one figure showing data.
The P. halocryophilus genome is made up of 35 contiguous strands, composed in a total of 3,428,475 base pairs, with a content of G+C base pairing at 40.5%. The genome is made up of 85.1% coding bases, of which 53.3% are coding positive strand and the other 46.7% code negative strand.
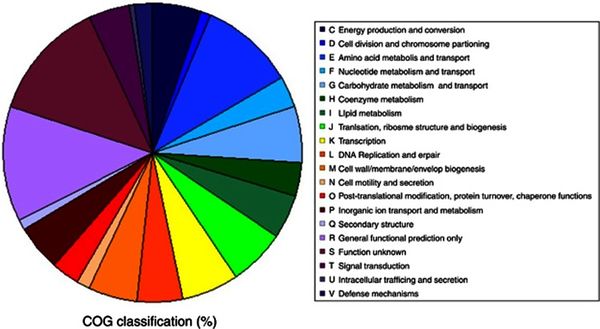
Figure 3 presents a pie chart for the distribution of genes in correspondence to specific functions. The orthologous groups presented that are related to cellular metabolism show a balanced distribution of about 5% of the total genome. The discernable exception comes from amino acid transport and metabolism, a functional category which makes up roughly 10% of the total genome, the largest orthologous group cluster accounted for with known function. The ratios from Figure 3 present closely comparable results to the genomic classifications of other cryophiles such as P. ingrahamii, E. sibiricum, and P. arcticus. [10]
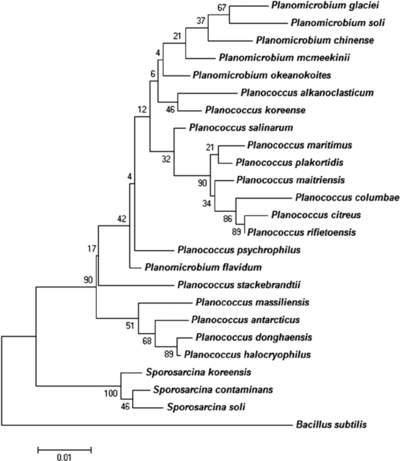
The phylogenetic position of P. halocryophilus was determined by a 16S rRNA gene sequence (amounting to 1550 base pairs) amplified by PCR. This determined that OR1 would be designated with the genus Planococcus and affiliated with the Planomicrobium clade. The P. halocryophilus strain’s location was identified by homologous signature nucleotides to the Planococcus genus, however the strain showed slight varying of clustering patterns compared to that of the Planomicrobium clade. The 16S rRNA genome sequence showed greatest relation to P. dorghaensis (99.6%) and P. antarcticus (98.8%) out of all the Planococcus species, and the greatest separation from P. columbae (96.4%). In order for two organisms to be defined as part of the same species, the threshold of comparison is designated as greater than or equal to 70% similarity by DNA-DNA comparison. Upon comparing DNA sequences of P. halocryophilus to that of P. dorghaensis, the similarity was measured at 46%, and for P. antarcticus at 18.2% similarity. These DNA similarity measurements provided concrete evidence that OR1 was an independent species from other Planococcus strains. [11]
Figure 4 shows a phylogenetic tree related to 16S RNA gene sequences among related taxa of Planococcus and Planomicrobium.
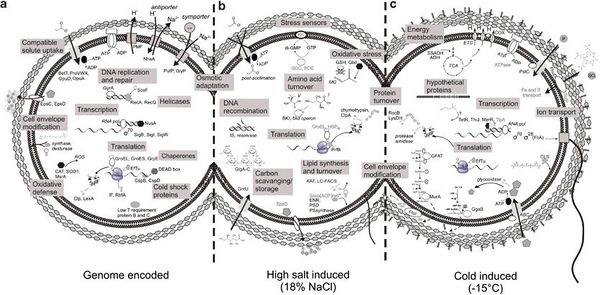
P. halocryophilus contain many genomic adaptations to both cold and osmotic stress through proteins that function in response to stress from cold temperatures, high salinity, and general stress. The wide array of proteins creates mechanisms of regulation and repair, act as chaperones, and function as shock proteins. These proteins are encoded by roughly 90 known loci of the OR1 strand genome. 43 loci account for proteins that respond to osmotic stress and adaptation to high salt conditions. Nine loci account for proteins that protect and repair translational factors. Eight loci account for proteins that function in membrane and cell envelope alteration. Seven loci account for proteins stat function as sigma factors. Six loci account for proteins that are involved with DNA replication and repair. Five loci account for cold shock proteins. Finally, there are 11 loci that account for proteins that respond to miscellaneous stress factors, one of which is an SOS-response repressor. These loci provide 90 examples of how cryophiles like P. halocryophilus are able to survive and thrive in harsh subzero saline environments. [12]
In order to maintain critical functions within transcription, translation, and regulation, P. halocryophilus contains specific cold-adapted proteins, observed in many cryophiles and mesophiles, that are able to provide essential functions under both cold and osmotic stress. In DNA/RNA replication, specialized helicases and recombination factors are dependable to drive replication in subzero conditions. Other specialized transcription and translation factor proteins have been observed to maintain functions at low temperature or in osmotic stress, including for transcription termination, translation initiation, elongation, and termination. As an alternative under extreme osmotic and subzero stress conditions, specialized sigma factors can be appointed to maintain signal transduction.
Figure 4 illustrates three segments of P. halocryophilus, (a) containing adaptive traits encoded by the genome, (b) representing adaptive mechanisms induced by osmotic stress, and (c) showing adaptive mechanisms induced under subzero stress. This figure functions as visual representation for only some of the many understood adaptive and synergistic traits and mechanisms that P. halocryophilus and other cryophiles must demonstrate and regulate in order to survive in high stress environments.[13]
Ecology
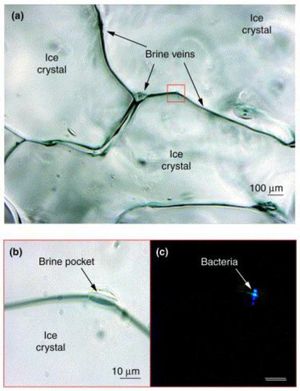
Planococcus halocryophilus is found in active surface layers of the Canadian high arctic permafrost (Eureka, Ellesmere Island), existing in brine veins between ice crystals, which reach subzero temperatures. These veins provide liquid water below freezing point on account of the high saline content that keeps the water from freezing. Liquid water channels act as critical accommodations for P. halocryophilus on account of their motile conduits for nutrients to reach the bacterial colonies. [14]
Figure 5 highlights the microscale habitat of P. halocryophilus through (a), an enhanced photo of bordering ice crystals, focused on the brine vein spaces in between. In (b), the photo is enlarged to reveal a brine pocket within the vein, a likely candidate for major microbiological activity, where (c) shows the same brine pocket but under epifluorescence to provide evidence of the inhabiting cryophilic bacteria. [15]
P. halocryophilus only accounts for a small fraction of the flourishing microbiota in arctic environments, and contributes to the large-scale emission of stored carbon dioxide that occurs alongside the melting of the permafrost. This disruption of the global carbon cycle has been shown to accelerate melting effects of global warming.
Astrobiological Significance
Astrobiology involves the study of life on Earth in order to predict the conditions of extraterrestrial life, ultimately to explore origins of life in the universe. The most important foundational question of astrobiology is what factors constitute habitat suitability, in other words, what must an environment require to be habitable. The most basic way to answer this question is to identify a presence of life; this provides a proxy for what makes that specific environment able to sustain life. In order to actually measure habitat suitability (to what degree is the environment suitable to life), three basic constituents must be taken into account. First, the environment in question; the physical, chemical and biological facets that make that environment unique. Second, the life form in question; how can the organism be classified and what components are distinctive of its ability to survive. Third, the relationship between the life and the environment that allow for the organism to grow, reproduce, and interact with that environment. [16]
P. halocryophilus plays a key role in Astrobiological research on account of its comparable habitat to that on Mars. Pockets of shallow ice have been discovered on the surface of Mars, and the permafrost habitats on Earth are used as comparable exemplary environments. The idea is that patterns of life in Earths harshest environments (of those that can be compared to environments on other planets and moons) are the best way to determine what traits life would need to survive in those environments, and accordingly, what degree of evolution would it take for similar life to possibly inhabit these extraterrestrial environments in question. From the brine veins that would exist between ice crystals on Mars’ surface, the most likely lifeforms to exist would need to contain many analogous traits to that of a cryophile such as P. halocryophilus. OR1 acts as the best test subject for the study of cryoenvironments because it is capable of growth at the lowest recorded temperatures, and is therefore the most extreme example of a cryophile.[17]
Proteomic Research
The study Raymond-Bouchard et al., 2017 conducted a proteomic analysis of Planococcus halocryophilus in order to identify key proteins involved in adaptive mechanisms for subzero growth and stress conditions for the extremophile. The study involved three growth conditions: a control at 23ºC, a subzero condition at -15ºC, and a halophilic condition at control temperature 23ºC and a NaCl content of 12%. Proteomic analysis of P. halocryophilus revealed notable expression under stress conditions of proteins in clusters of orthologous groups and KEGG pathways involving cell wall synthesis and modification, fatty acid metabolism, transportation, translation processes, amino acid/nucleotide turnover and metabolism, DNA replication/repair and transcription, oxidative stress, and energy acquisition. [18]
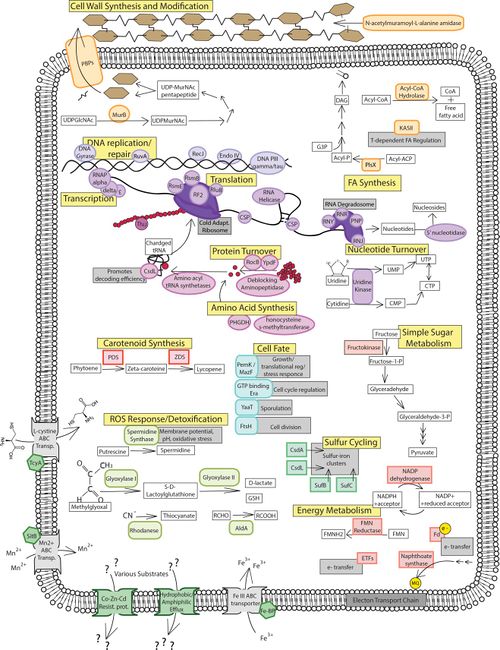
Some proteins involved in stabilizing transcription and translation processes in cold conditions include cold-shock proteins, RNA helicase, and protein chaperones such as ThiJ. These proteins are found to be abundant in all three conditions (control temp, low temp, and high salt), which suggests that P. halocryophilus is prepared to react quickly to temperature fluctuation. ThiJ is an important protein for oxidative stress response in cold conditions and functions as a protein deglycase, tripling in abundance under low temperature conditions. [19]
Proteins associated with cell wall synthesis and modification include DD-transpeptidase and MurB. DD-transpeptidases are in charge of peptide chain cross-linkages in cell wall synthesis, and is increased in abundance by nine-fold under subzero conditions. MurB plays an enzymatic role in peptidoglycan synthesis and is more than doubly expressed under subzero conditions. These proteins can be referenced in figure 6 under the cell wall synthesis and modification label. Increased peptidoglycan synthesis plays a crucial role in P. halocryophilus adaptation, as it creates negative charge in the matrix, enabling CaCO3 to form the encrustations observed on the cell wall during subzero growth, providing extra protection to the cell. [20]
One of the proteins concerned with fatty acid synthesis was a phosphate called acyl-ACP acyltransferase (PlsX). The PlsX phosphate provides a linkage between the fatty acid synthase and phospholipid synthesis pathways, and is increase by nine-fold in sub-zero, however is not observed in the other two conditions. It plays a critical role in modulating the two synthesis pathways, as well as unsaturated fatty acid accumulation in the cell membrane. This is contradictory to findings discussed earlier (in Mykytczuk et al., 2013) where saturated fatty acids were abundant in the cell membrane in subzero temperatures in place of unsaturated, which was the other way around for most psychrophiles and makes functional sense on account of increased membrane fluidity from long chain UFAs. Another way to modulate membrane fluidity in psychrophiles is through carotenoid pigments. Proteins engaged in the carotenoid (lycopene) synthesis include two enzymes, carotene desaturase and phytoene desaturase, which were most abundant at subzero and halophilic stress conditions. Carotenoids manipulate fatty acid composition in the membrane to prefer branched chain UFAs, increasing membrane rigidity as well as balanced fluidity. While this function also contradicts the findings (of Mykytczuk et al., 2013), further research must be conducted in order to understand the role of these abundances. The proteins for both fatty acid and carotenoid synthesis can be referenced in figure under their respective labels. [21]
Psychrophiles must develop membrane adaptations to overcome lower transport and diffusion rates in cold environments, for which transporter proteins are increased heavily in abundance. P. halocryophilus contains three ABC transporter binding proteins that are incorporated in the transport of cysteine, manganese, and ferric iron transport respectively. All three ABC transporter binding proteins had increased in abundance in subzero conditions as well as halophilic conditions for the cysteine and ferric iron binding proteins. [22]
Proteins connected to translation and ribosomal processes include CsdL, peptide chain release factor 2, pseudouridine synthase B, RsmB, RsmE, GTP binding proteins, and S17. CsdL is responsible for identifying noncognate codons and sustaining efficiency in decoding by analyzing adenine in tRNA, and is present in the two stress conditions. Peptide chain release factor 2 is an enzyme that releases complete peptide chains by attaching to the ribosome to end mRNA translation via the stop codon, and is present only in subzero conditions. Pseudouridine synthase B is a ribosomal large subunit implicated as an RNA chaperone and in the initiation of translation, increasing five-fold in abundance under subzero conditions.
RsmB and RsmE are both rRNA small subunit methyltransferases, involved in rRNA stabilization and translation regulation, quadrupling and doubling in abundance respectfully for subzero conditions. GTP binding proteins are incorporated into ribosome biogenesis and see increased abundance at subzero conditions. S17 is a 30S ribosomal protein involved in regulating accuracy of translation, and is a unique example of a ribosomal protein that is reduced in abundance under stress conditions in favor of other ribosomal functions. This is due to the preference of increasing translation rates at -15ºC conditions, however translation must be supervised to some extent in order to avoid error while maintaining efficiency. Translation is the theoretical rate-limiting step for protein synthesis of psychrophiles, on account of diffusion rate reduction in subzero conditions. Translation proteins and ribosomal processes are referenced in figure 6, under the translation label. [23]
Proteins associated with amino acid synthesis include uridine kinase, homocysteine S-methyltransferase, chorismate synthase, D-3-phosphoglycerate dehydrogenase, and arginase. Uridine kinase is an enzyme that participates in the nucleotide salvage pathway and is only expressed under subzero conditions. Homocysteine S-methyltransferase is an enzyme complex that catalyzes methionine production from the amino acid homocysteine, and is most abundant in -10ºC conditions. Chorismate synthase and D-3-phosphoglycerate dehydrogenase are both involved in amino acid synthesis and increase expression under both subzero and halophilic conditions. Arginase is an enzyme that produces ornithine by breaking down arginine, and shows a decrease in expression under stress conditions. [24]
Proteins engaged in DNA replication and repair and well as transcription include endonuclease IV, DNA gyrase B, RNAP∑, and MtrB. Endonuclease IV is an enzyme that identifies and primes damaged cites for DNA repair, and tripled inexpression under stress conditions. DNA gyrase B functions in response to DNA supercoiling that occurs from stress conditions, and mediates the repair and recovery of the nucleic acid material.
RNAP∑ is a subunit of the RNA polymerase enzyme and is abundant under cold conditions and absent in control, with a stress response function that is largely unknown.
MtrB is a unique transcription factor used in transcription attenuation involved with the Trp operon, and is an exception to gene expression control because its abundance is increased under stress conditions. Proteins involved in ROS response and detoxification include spermidine synthase, and AldA. Spermidine synthase is the enzyme that produces the polyamine spermidine, which reduces ROS DNA strand damage by scavenging free radicals. AldA is an aldehyde dehydrogenase that lyses toxic products of oxidative stress including glyoxal and methylglyoxal. ROS detoxification requires the removal of adjunct RES from which carbonyls can be recycled and used under stress conditions in metabolism. [25]
Proteins connected to energy metabolism include phosphoglycerate mutase, UTP-glucose-1-phosphate uridylyltransferase, ATP synthase, FMN reductase, glutamate-1-semialdehyde aminotransferase, ferredoxin, NADPH dehydrogenase, naphthoate synthase, Ccs1/ResB, fructokinase, and CsdA/SufS. Phosphoglycerate mutase, UTP-glucose-1-phosphate uridylyltransferase, and ATP synthase are all proteins that decrease in abundance in response to cold-stress conditions. They are proteins associated with glycolysis, glucose metabolism, and ATP synthesis respectively. FMN reductase, glutamate-1-semialdehyde aminotransferase, ferredoxin, NADPH, dehydrogenase, naphthoate synthase, and Ccs1/ResB are proteins that increase in abundance in response to cold-stress conditions. Fructokinase is an enzyme that breaks down the simple sugar fructose as an alternative to glucose metabolism and is abundant as a general stress response because fructose is a metabolite that is tolerant to stress conditions. CsdA/SufS is a sulfur source protein within P. halocryophilus, donating sulfur in the process of sulfur cycling. [26]
Proteins associated with cell growth include PemK/MazF, Era, YaaT, FtsH, and FtsZ. PemK/MazF is a transcriptional regulator that is only detectable at subzero conditions. Era is a GTP-binding protein participating in cell cycle regulation, assembly of ribosomes, and metabolism that increased five-fold in the -10ºC condition. YaaT is a cell fate regulator that tripled in abundance at subzero. FtsH and FtsZ are both cell division proteins that increase in abundance at subzero conditions. Era, Yaat, FtsH, and FtsZ are also increased in expression under halophilic conditions, suggesting that they function as general stress response rather than specifically to cold conditions. These proteins are examples of stress responses that limit growth under energetically favorable circumstance in priority of survival over growth. [27]
From the proteomic data between -10ºC and 23ºC + NaCl conditions, prominent differences in abundance were noted from proteins involved in peptidoglycan biosynthesis, DNA replication and repair, as well as amino acid, sulfur, and B6 metabolisms. Due to greater abundance from these proteins in subzero conditions over halophilic conditions, these processes are held to be important for subzero growth specifically. [28]
References
- ↑ [Bacterium Planococcus Halocryophilus Offers Clues about Microbial Life on Enceladus, Mars. www.sci-news.com/space/article01105-planococcus-halocryophilus-bacterium.html.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Planococcus halocryophilus Sp. Nov., an Extreme Sub-Zero Species from High Arctic Permafrost.” International Journal of Systematic and Evolutionary Microbiology, Microbiology Society, 1 Aug. 2012, ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.035782-0.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Bacterial Growth at -15 ºC: Molecular Insights from the Permafrost Bacterium Planococcus halocryophilus Or1.” Nature News, Nature Publishing Group, 7 Feb. 2013, https://www.ncbi.nlm.nih.gov/pubmed/23389107.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Planococcus halocryophilus Sp. Nov., an Extreme Sub-Zero Species from High Arctic Permafrost.” International Journal of Systematic and Evolutionary Microbiology, Microbiology Society, 1 Aug. 2012, ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.035782-0.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Bacterial Growth at -15 ºC: Molecular Insights from the Permafrost Bacterium Planococcus halocryophilus Or1.” Nature News, Nature Publishing Group, 7 Feb. 2013, https://www.ncbi.nlm.nih.gov/pubmed/23389107.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Planococcus halocryophilus Sp. Nov., an Extreme Sub-Zero Species from High Arctic Permafrost.” International Journal of Systematic and Evolutionary Microbiology, Microbiology Society, 1 Aug. 2012, ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.035782-0.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Bacterial Growth at -15 ºC: Molecular Insights from the Permafrost Bacterium Planococcus halocryophilus Or1.” Nature News, Nature Publishing Group, 7 Feb. 2013, https://www.ncbi.nlm.nih.gov/pubmed/23389107.]
- ↑ [Diomande, et al. “Role of Fatty Acids in Bacillus Environmental Adaptation.” Frontiers, 23 July 2015, www.frontiersin.org/articles/10.3389/fmicb.2015.00813/full.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Bacterial Growth at -15 ºC: Molecular Insights from the Permafrost Bacterium Planococcus halocryophilus Or1.” Nature News, Nature Publishing Group, 7 Feb. 2013, https://www.ncbi.nlm.nih.gov/pubmed/23389107.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Bacterial Growth at -15 ºC: Molecular Insights from the Permafrost Bacterium Planococcus halocryophilus Or1.” Nature News, Nature Publishing Group, 7 Feb. 2013, https://www.ncbi.nlm.nih.gov/pubmed/23389107.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Planococcus halocryophilus Sp. Nov., an Extreme Sub-Zero Species from High Arctic Permafrost.” International Journal of Systematic and Evolutionary Microbiology, Microbiology Society, 1 Aug. 2012, ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.035782-0.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Bacterial Growth at -15 ºC: Molecular Insights from the Permafrost Bacterium Planococcus halocryophilus Or1.” Nature News, Nature Publishing Group, 7 Feb. 2013, https://www.ncbi.nlm.nih.gov/pubmed/23389107.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Bacterial Growth at -15 ºC: Molecular Insights from the Permafrost Bacterium Planococcus halocryophilus Or1.” Nature News, Nature Publishing Group, 7 Feb. 2013, https://www.ncbi.nlm.nih.gov/pubmed/23389107.]
- ↑ [Mykytczuk, Nadia C. S., et al. “Planococcus halocryophilus Sp. Nov., an Extreme Sub-Zero Species from High Arctic Permafrost.” International Journal of Systematic and Evolutionary Microbiology, Microbiology Society, 1 Aug. 2012, ijs.microbiologyresearch.org/content/journal/ijsem/10.1099/ijs.0.035782-0.]
- ↑ [Feller, G. “Cryosphere and Psychrophiles: Insights into a Cold Origin of Life” Life (Basel, Switzerland)., U.S> National Library of Medicine, 11 June 2017, www.ncbi.nlm.nih.gov/pubmed/28604605.]
- ↑ [Cockell, C S, et al. “Habitability: A Review.” Astrobiology., U.S. National Library of Medicine, Jan. 2016, www.ncbi.nlm.nih.gov/pubmed/26741054.]
- ↑ [Cockell, C S, et al. “Habitability: A Review.” Astrobiology., U.S. National Library of Medicine, Jan. 2016, www.ncbi.nlm.nih.gov/pubmed/26741054.]
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
- ↑ Raymond‐Bouchard, Isabelle, et al. “Mechanisms of Subzero Growth in the Cryophile Planococcus halocryophilus Determined through Proteomic Analysis.” Freshwater Biology, Wiley/Blackwell (10.1111), 13 Oct. 2017, onlinelibrary.wiley.com/doi/epdf/10.1111/1462-2920.13893.
Authored for BIOL 238 Microbiology, taught by Joan Slonczewski, 2018, Kenyon College.