Pseudomonas and Bioremediation
According to the Center for Climate and Energy Solutions, petroleum supplies 32.5% of global energy use. 1 The global drilling and transport of petroleum has resulted in oil spills in marine ecosystems, such as the highly publicized Deepwater Horizon oil spill off the Gulf of Mexico, the Prestige oil spill in Spain, and the Exxon Valdez oil spill in Alaska. Oil spills are costly and ecologically damaging as polycyclic aromatic hydrocarbons have been linked to cardiac arrhythmia in marine organisms. 2 There is a great need for efficient and ecologically friendly remediation strategies.
Bioremediation is a strategy that uses naturally occurring organisms to break down pollutants, such as petroleum hydrocarbons, into less toxic substances. Species in the bacterial genus Pseudomonas present high potential for hydrocarbon degradation due to their metabolic diversity, their abundance in microbial communities, and their resistance to chemical remediation agents present at contamination sites. 3
Psuedomonads
Classification
Domain: Bacteria
Phylum: Proteobacteria
Class: Gammaproteobacteria
Order: Pseudomonadales
Family: Pseudomonadaceae
Genus: Pseudomonas4
General Characteristics
Pseudomonas is a genus of Gram-negative, rod-shaped bacteria. They are aerobic and non-sporulating with one or more polar flagella for motility. 3 There are currently 218 species assigned to Pseudomonas and the genus has considerable heterogeneity.5 Pseudomonas contains a number of scientifically and medically studied bacteria, such as the opportunistic pathogen P. aeruginosa, and there is increasing availability of Pseudomonas strain genome sequences. 6 Pseudomonas is well known for its metabolic versatility, being able to utilize an unusually wide range of organic compounds. 7 Certain members of the genus are able to metabolize pollutants, including P. fluorescens,8 P. putida, P. cepacia, P. vesicularis, and P. paucimobilis .9 As a result, they are often isolated and studied for their bioremediation capabilities.
The metabolic versatility of the genus allows Pseudomonas species to inhabit a wide variety of environments. They are widespread in nature, inhabiting soil and marine habitats as well as plant and animal tissue. 10 Pseudomonas species have been found to dominate hydrocarbon degrading bacterial communities even in the Artic—an environment that entails extreme temperature conditions, limited availability of nutrients such as nitrogen, and low levels of available water. 11
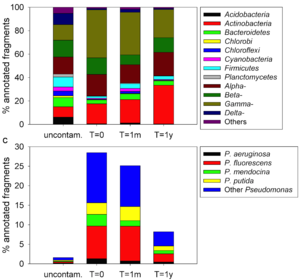
In a study that sequenced the metagenome of contaminated and uncontaminated Artic soil, researchers found that Gammoproteobacteria comprised up to 30% of the microbial community at t=0 (Figure 1A). Metagenomic sequencing revealed that the dominance of Gammoproteobacteria was mainly due to Pseudomonas species (Figure 1B). Consistent with its relative abundance, researchers confirmed with PCR that Pseudomonas was actively expressing alkane hydroxylase genes and was responsible for up to 20% of total gene expression. Pseudomonas abundance dropped off after a year, at t = 1y, after prolonged degradation of available hydrocarbons.
Response to Oil Contamination
Pseudomonas has shown robust response to oil contamination both in vitro and in situ. A 2013 study found that Pseudomonas was able to respond to new oil contamination by increasing its abundance and changing its community structure.12 Microbial community differentiation was observed in the Pseudomonas genus in response to oil contamination.12
In a metagenomic analysis of the microbial population in petroleum pipelines, Pseudomonas stutzeri was detected as the most abundant hydrocarbon degrading organism.13 Pseudomonas stutzeri’s relative abundance was seen as a result of its ability to utilize aromatic hydrocarbons such as xylene, toluene, phenol, as well as polycyclic aromatic hydrocarbons, such as naphthalene. Researchers were able to amplify the genes for the enzymes used in the degradation of hydrocarbons from Pseudomonas stutzeri. These enzymes include: benzoate dioxygenase, toluene 1,2-dioxygenase, and catechol 1,2-dioxygenase, which play a role in aromatic hydrocarbon degradation, and methanol dehydrogenase, which is involved in aliphatic hydrocarbon degradation.13
Metabolism
Petroleum Hydrocarbons
Petroleum hydrocarbons are a major toxic contaminant in the marine environment. Two to ten million tons of crude petroleum oil are spilled annually into marine environments.14 The biodegradability of petroleum components generally decreases in the following order: n-alkanes, branched-chain alkanes, branched alkenes, low molecular weight ¬n-alkyl aromatics, monoaromatics, cyclic alkanes, polycyclic aromatic hydrocarbons.14 Pseudomonas strains are capable of degrading petroleum hydrocarbons and aromatic hydrocarbons such as benzene, toluene, ethylbenzene, and xylene.9
Polycyclic aromatic hydrocarbons
Polycyclic aromatic hydrocarbons (PAHs), organic chemicals consisting of three or more benzene rings, have the lowest biodegradability out of all petroleum components. They are especially problematic because of their carcinogenic potential and their tendency to persist in an environment due to their low water solubility.15 The Environmental Protection Agency (EPA) has listed 16 PAHs as priority pollutants.16 A 2014 study found that the elevated PAH concentrations after the Deepwater Horizon oil spill, which spilled 636 million liters of crude oil into the Gulf of Mexico, negatively impacted the cardiac function of resident fish species.2 Humans can ingest PAHs through cooked foods and the EPA has listed seven PAHs as probable human carcinogens.17
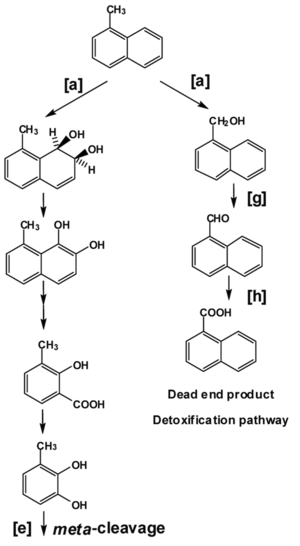
In addition to metabolizing more easily degradable petroleum components such as aromatic hydrocarbons, Pseudomonas strains also have the metabolic potential to degrade PAHs such as naphthalene, acenaphthene, anthracene, fluoranthene, and phenanthrene.9 Their ability to readily degrade recalcitrant PAHs gives Pseudomonas species high bioremediation potential. The metabolism of naphthalene by Pseudomonas putida can be used to exemplify Pseudomonas PAH catabolism. Pseudomonas putida uses dioxygenase enzymes (naphthalene dioxygenase, Figure 2 [a]) to catabolize the formation of cis-1,2dihydrodiol from naphthalene. Not shown in Figure 2, an iron-sulfur flavoprotein reductase accepts electrons from NADPH and transfers them to a ferredoxin. Ferredoxin reduces the terminal iron-sulfur protein which catabolizes the oxidation of naphthalene to form naphthalene cis-1,2dihydrodiol (seen in Figure 2, as the first compound after [a]).9 This diol is further oxidized by dehydrogenase enzymes to catechol, which enters the tricarboxylic acid cycle after meta-cleavage (Figure 2 [e]). Pseudomonas putida is capable of utilizing naphthalene as its sole source of carbon and energy through this metabolic pathway. Alternatively, it can hydroxylate the side-chain, leading to the eventual detoxification of naphthalene (Figure 2 [h]).18
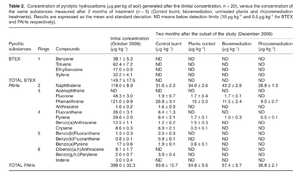
The metabolic capability of Pseudomonas putida to degrade naphthalene is exhibited in the data represented in Figure 3. The study used burnt soil, which contains a high concentration of PAHs, to study Pseudomonas putida bioremediation capability. After two months of bioremediation treatment with Pseudomonas putida strains, the concentration of naphthalene decreased by 63.6%.19 Pseudomonas putida was also effective at degrading pyrene, a high-molecular weight PAH, and reduced the total amount of pyrene by 96.6%. High-molecular weight PAHs, such as pyrene, are more recalcitrant and possess even lower biodegradability than other petroleum components—once again demonstrating the remediation potential of Pseudomonas species.20
Multiple studies affirm the robust metabolism of Pseudomonas. In a different study, researchers found similar results when they inoculated bottles of toluene, octane, and naphthalene with Pseudomonas strains. At 25 degrees Celsius, the compound naphthalene was undetectable after 1 to 2 days.21
Research Applications
Genetic Bioaugmentation
The metabolic versatility of Pseudomonas is reflected in its large genome size, typically above 6 Mb. Horizontal gene transfer via plasmids, phages, and transposons has been reported in Pseudomonas, which aids in the distribution of the genus’s diverse metabolic pathways. The metabolic capacities of Pseudomonas can be further enhanced and exploited through genetic bioaugmentation in order to develop more efficient bioremediation strategies.
Pseudomonas putida exhibits the metabolic capability to degrade naphthalene, a recalcitrant high-molecular weight PAH. The genes for naphthalene dioxygenase from Pseudomonas putida are encoded on the NAH7 plasmid and the nucleotide sequence for the genes is publicly available.22 Furthermore, researchers have cloned and shown the expression of naphthalene dioxygenase activity in E. coli. E. coli containing plasmids carrying the dioxygenase gene were studied for their ability to express the gene and produce dihydrodiol from naphthalene. Using liquid chromatography and nuclear magnetic resonance, the study observed dihydrodiol yields ranging from 50 to 94%23 The successful transfer of naphthalene degradation ability shows the potential of genetic bioaugmentation strategies in Pseudomonas.
Compatibility with Chemical Remediation
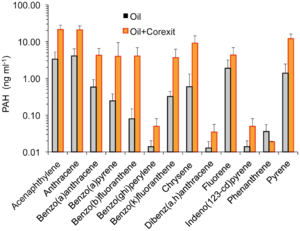
Dispersants are commonly used spill-treating agents that promote the formation of smaller oil droplets. This increases the surface area to volume ratio of the oil droplet and allows a greater number of hydrocarbon degrading bacteria to come into direct contact with the oil. A study that investigated the impact of dispersants after the Deepwater Horizon oil spill found that dispersants increased the mobility of PAHs, decreasing the persistence of PAHs in the marine environment.24 The dispersant Corexit increased the availability of PAHs by approximately 10 fold for all PAHs measured, except phenanthrene (Figure 4).24
Dispersants increase the efficiency of cleaning, but due to their toxicity, it is important to evaluate their influence on the biodegradation rate of bacteria. In a study to evaluate the inhibitory effects on the growth of Pseudomonas species, the experimental results revealed that dispersants were not significantly toxic to Pseudomonas.25 As gram-negative bacteria, Pseudomonas species are less sensitive to spill treating agents than gram-positive bacteria.25 The negligible sensitivity of Pseudomonas species to the dispersants tested in the study make it an ideal organism to accompany certain types of chemical remediation.
Further Reading
Although the study referenced above showed no inhibitory effects of dispersants, the results were obtained from in vitro studies, so the effects of spill treating agents should be further assessed on the evolution of a bacterial community in seawater.25 There is a need for greater research on the vitality of microbe communities in the presence of dispersants in order to determine if chemical remediation can be effectively used in combination with bioremediation. Current literature is divided. The study below shows that while dispersants unambiguously enhanced the bioavailability of PAHs, certain dispersants inhibited the growth of P. putida, counteracting the effect of the increased bioavailability.
Biostimulation is another promising strategy that can be used to enhance the bioremediation process. Biostimulation investigates the nutrients that are required to support microbial activity and attempts to stimulate biodegradation rates through the addition of nutrients. The study below carries out experiments with three different nitrogen supplementations.
Bioremediation potential of microorganisms from a sandy beach affected by a major oil spill
More research:
Review paper of bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes
A research paper studying the effects of chemical remediation in conjunction with bioremediation
References
1 Center for Climate and Energy Solutions
2 Incardona et al., 2014, J. Incardona, L.D. Gardner, T.L. Linbo, T.L. Swarts, A.J. Esbaugh, E.M. Mager, et al. Deepwater Horizon crude oil toxicity to the developing hearts of large predatory pelagic fish. Proc Natl Acad Sci U S A, 111 (2014), pp. E1510–E1518
3 Özen, A. I., & Ussery, D. W. (2012). Defining the Pseudomonas Genus: Where Do We Draw the Line with Azotobacter? Microbial Ecology, 63(2), 239–248. doi:10.1007/s00248-011-9914-8
4 NCBI Taxonomy Database
5 List of prokaryotic names with standing in nomenclature
6 Pseudomonas Genome Database
7 Stanier RY, Palleroni NJ, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271.
8 Pavel Kuráň, Josef Trögl, Jana Nováková, et al., “Biodegradation of Spilled Diesel Fuel in Agricultural Soil: Effect of Humates, Zeolite, and Bioaugmentation,” The Scientific World Journal, vol. 2014, Article ID 642427, 8 pages, 2014. doi:10.1155/2014/642427
9 Cerniglia, C. E.Biodegradation of polycyclic aromatic hydrocarbons Biodegradation 1992, 3,351– 368
10 Joint Genome Institute, U.S. Department of Energy
11 Yergeau E, Sanschagrin S, Beaumier D, Greer CW (2012) Metagenomic Analysis of the Bioremediation of Diesel-Contaminated Canadian High Arctic Soils.PLoS ONE 7(1): e30058. doi: 10.1371/journal.pone.0030058
12 Reis, I., Almeida, C. M. R., Magalhaes, C. M., Cochofel, J., & Guedes, P. (03/2014). Environmental science and pollution research international: Bioremediation potential of microorganisms from a sandy beach affected by a major oil spill Ecomed. doi:10.1007/s11356-013-2365-7
13 Joshi, M., Dhebar, S. V., Dhebar, S. V., Bhargava, P., & Pandit, A. (08/2014). Archives of microbiology: Metagenomics of petroleum muck: Revealing microbial diversity and depicting microbial syntrophy Springer. doi:10.1007/s00203-014-0992-0
14 Tyagi M, da Fonseca MMR, de Carvalho CC (2011) Bioaugmentation and biostimulation strategies to improve the effectiveness of bioremediation processes. Biodegrad 22:231–241
15Polycyclic Aromatic Hydrocarbons, Environmental Protection Agency
16 J., Wang, L., Fu, P. P., & Yu, H. (2004). Photomutagenicity of 16 polycyclic aromatic hydrocarbons from the US EPA priority pollutant list.Mutation Research, 557(1), 99–108. doi:10.1016/j.mrgentox.2003.10.004
17 Polycyclic organic matter, Environmental Protection Agency
18 V. Paliwal, S.C. Raju, A. Modak, P.S. Phale. H.J. Purohit. Pseudomonas putida CSV86: A Candidate Genome for Genetic Bioaugmentation. PLOS One. DOI: 10.1371/journal.pone.0084000. 2014
19 Pizarro-Tobías, P., Fernández, M., Niqui, J. L., Solano, J., Duque, E., Ramos, J.-L. and Roca, A. (2015), Restoration of a Mediterranean forest after a fire: bioremediation and rhizoremediation field-scale trial. Microbial Biotechnology, 8: 77–92. doi: 10.1111/1751-7915.12138
20 M.M. O’Mahony, A. Dobson, J.D. Barnes, I. Singleton. The use of ozone in the remediation of polycyclic aromatic hydrocarbon contaminated soil. Chemosphere, 63 (2006), pp. 307–314
21 Whyte, L. G., Bourbonniére, L., & Greer, C. W. (1997). Biodegradation of petroleum hydrocarbons by psychrotrophic Pseudomonas strains possessing both alkane (alk) and naphthalene (nah) catabolic pathways. Applied and Environmental Microbiology, 63(9), 3719–3723.
22 Simon MJ, Osslund TD, Saunders R, Ensley BD, Suen W-C,Cruden DL, Gibson DT & Zylstra GJ (1992) Nucleotide sequences encoding the genes for naphthalene dioxygenase in Pseudomonas putida G7 and Pseudomonas sp. NCIB 9816--4. Gene (in press)
23 P. Di Gennaro, E. Galli, G. Albini, F. Pelizzoni, G. Sello, G. Bestetti, Production of substituted naphthalene dihydrodiols by engineered Escherichia coli containing the cloned naphthalene 1,2-dioxygenase gene from Pseudomonas fluorescens N3, Research in Microbiology, Volume 148, Issue 4, 1997, Pages 355-364, ISSN 0923-2508
24 Zuijdgeest A, Huettel M (2012) Dispersants as Used in Response to the MC252-Spill Lead to Higher Mobility of Polycyclic Aromatic Hydrocarbons in Oil-Contaminated Gulf of Mexico Sand. PLoS ONE 7(11): e50549. doi: 10.1371/journal.pone.0050549
25 Diego Rial, Miguel A. Murado, Ricardo Beiras, José A. Vázquez, Toxicity of four spill-treating agents on bacterial growth and sea urchin embryogenesis, Chemosphere, Volume 104, June 2014, Pages 57-62, ISSN 0045-6535
Edited by Jessica Cheng, a student of Suzanne Kern in BIOL168L (Microbiology) in The Keck Science Department of the Claremont Colleges Spring 2015.
