Swarming Motility in Proteus Mirabilis: Causative Agent of UTIs
Introduction
By Nicole Valentini
Proteus mirabilis belongs to the family Enterobacteriacae, which are gram negative, facultatively anaerobic rods that have the ability to grow in nutrient deficient environments (28). Many species in the family are highly motile, with numerous flagella that allow for several different modes of locomotion. Enterobacteriacae are also known to cause many diseases in both plants and animals. One example is the Erwinia species, which causes defects such as wilts and galls in an array of plants (1).
P. mirabilis is normally found in the human intestine along with other organisms composing a highly complex micro flora. They also inhabit other outside environments, and are especially prevalent in hospitals and care facilities. Interestingly, P. mirabilis have been known to inhabit the skin and mucous of both patients and personnel working in these environments, which may be the primary vectors for pathogenecity.(2) Metabolically, P. mirabilis is involved in urease production, which is then used to convert urea to ammonia in the following reaction: (NH2)2CO -> 2NH3 + CO2. P. This may be one of the reasons the pathogen is so successful in colonizing the urinary tract and cause infection in humans.
Motility in P. mirabilis is highly complex as they engage in several different kinds of movement depending on the specific environment they are inhabiting. Most of these movements are directly tied to the differential expression of flagella and other factors. When in liquid environments, normal movement is facilitated by swimming. However, in more viscous and solid environments,P. mirabilis have the ability to differentiate in elongated, multinucleated, highly flagellated cells, which then allows them to move together over solid surfaces at very high rates.(3) This activity, known as swarming, is a primary factor in the success of P. mirabilis in causing complicated uriniary tract infections and other more serious bladder and kidney infections. (9)
UTIs due to f P. mirabilis are usually a secondary result of long-term catheterization in hospitals, or with individuals who have urinary structural abnormalities. (9) The bacteria’s ability to swarm over surfaces allows them to ascend up the urethra, eventually invading the bladder and kidneys. (10) P. mirabilis infection then leads to more complicated problems, such as bladder/kidney stones. In rare cases, P. mirabilis is able to enter the blood stream inducing a systemic inflammatory response syndrome (SIRS), which as a mortality rate of 20%-50%. (11)
In addition to their adaptive mobile abilities, other virulence factors have deemed P. mirabilis successful UTI causative agents. Overtime, their inhabitance in hospitals has led to the expression of several antibiotic resistance genes, making catheter associated urinary tract infections very difficult to treat. Furthermore, their ability to forms stones in the organisms bladder/kidneys provide the bacteria with a safe haven that is impenetrable to antibiotics or the host individuals immune defenses. (9) Several methods have been developed to prevent CAUTI infection, however treating them is much more complex, as they are resistant to many of the most common types of antibiotics (penicillin, cephalosporin, tetracycline, etc.). Current research is looking into other possible routes of treating CAUTIS in an effective and simple manner, and targets the cell interesting swarming ability and other virulence factors. Understanding P. mirabilis ability in conjunction with other virulence factors may lead to greater advances in treating CAUTIs and reducing it’s prevalence in hospital settings.
Swarming Motility: A Locomotive Advantage
The ability to swarm over more viscous or even solid surfaces is restricted to only a few bacterial families: Furmicutes, alpha proteobacteia, and gama proteobacteria. P. mirabilis falls under the gamma proteobacteria, which also include E. coli</> and Salmonella enterica, known to cause infection in other areas of the body. These organisms, especially E. coli, have been used as models in the laboratory in understanding the mechanisms directly related to the swarming motility of bacterial colonies. (2)
Generally, vegetative bacterial cells are engaged in what is known as swimming motility, which employs the use of a rotating flagellum. (2) An individual flagellum can be thought of as a motor, that is coupled to the flow of protons across the membrane to provide energy for rotation. The motor itself is made up of two rings connect to a helical filament. (12) The direction of rotation determines the direction in which the cell swims, and is regulated by sensory inputs tied to outside environmental influence. (12). Swimming is the primary locomotive method for P. mirabilis in liquid environments, and it differs in many ways to the mysterious swarming some in bacteria.
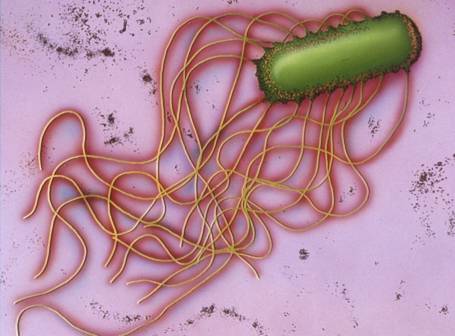
Swarming motility is mediated by several factors. The presence of a solid surface or highly viscous environments is necessary for this type of motility to occur, however it is still a mystery as to how solid surfaces may induce the several structural changes in a swarming bacterium. One of the most important changes to occur is the substantial increase of flagella along the cell surface. Bacterial cells become hyperflagellated with individual flagella arranged in bundles randomly along the surface. The bundle arrangement has been linked to increased stiffness and generation of more force to propel the cells in the necessary direction.(13) A recent study by Tuson et. al. 2012 was conducted using P. mirabilis to evaluate the effectiveness of increasing flagellum density in swarming. Interestingly, Tuson et. al. 2012 found that the increased surface density of flagellum (5 times greater than that of vegetative cells) produces an increase in cell velocity when travelling through viscous medium. (14) In addition to an increase in flagellum, swarmer cells become elongated and multinucleated, although these two components were not directly linked to an increase in velocity.(2,14) Some swarming bacterium secrete what is known as a surfactants, which forms a layer over the solid surface to facilitate easier movement. This layer is usually seen as a watery substance that is found slightly before the cell front, as was observed in several studies of Bacillus subtilis, another common swarmer cell. (15) Interestingly, P. mirabilis has not been found to be a sufacant producing swarmer cell, and in fact experiences an inhibitory response when in the presence of surfactants. (16)
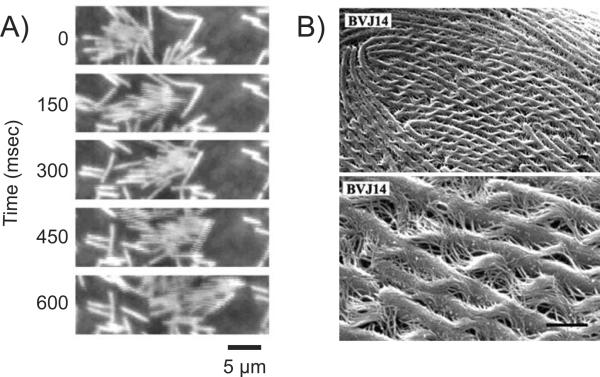
While swimming is possible on an individual cell basis, swarming is directed by the presence of multiple cell bodies and is facilitated as colonial movements. Swarmer cells come together and form “rafts” which are like side by side groups, solely linked by the interaction of flagella (Figure 1). Research is still being conducted to determine the necessity of raft formation in regards to swarming, however rafting is constantly observed in all bacterial swarmers, including P. mirabilis swarming over a catheter in CAUTI patients. (2)
Observing Swarming in the Laboratory
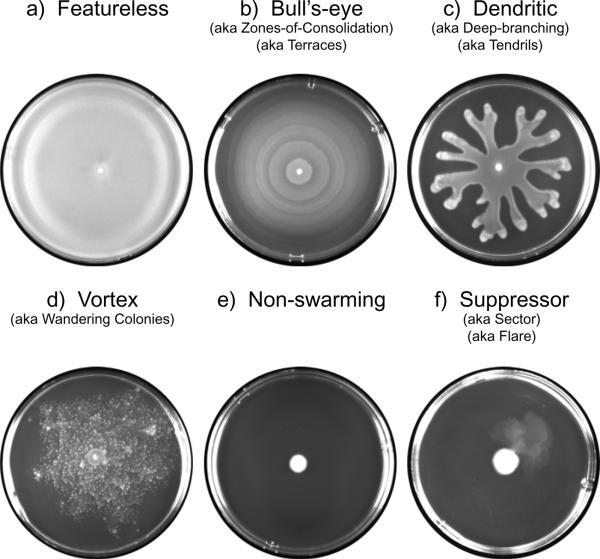
Swarming of different types of bacterial cells cultured on agar in a laboratory produces many distinct colonization patterns, and allows for a better visual understanding of the overall colonial structure of swarming bacteria. Swarming bacteria can produce a range of patterns, and that these patterns may be linked to different environmental conditions. (19) The most well-known and irregular swarming pattern in that formed by P. mirabilis. On solid agar plates, P. mirabilis grow outward in a “bull’s eye pattern.” This pattern is generated by waves of swarmer cell movement, followed by differentiation into vegetative cells, and then differentiation again back into swarmer cells. Each swarming cycle/differentiation results in the formation of a terrace, and thus the visual bull’s eye pattern. However, differentiation back into vegetative cells is not a necessary component of the bull’s eye pattern, as it was observed that Proteus vulgarus cause the same colonial plate pattern while consistently remaining swarmer cells. (20)
Other patterns of colonial swarmer cell formation are equally interesting. Dendrite formation occurs when swarmer colonies form long thing regions emulating out from the center of inoculation on a plate. This type of pattern has been observed in Proteus aeruginosa and B. subtilitis. Recent research has indicate that the dendrite formation may be a result of multiple surfcant secretion (21). Another interesting colonial formation is that of vortices, or wandering colonies that travel in radiating packs. Studies involving Panibacillus vortices indicate that swarming motility combined with a curved cell morphology may result in this type of pattern. Swarming can also produce no pattern, in which the swarms move continuously out from the center of the plate, making an almost transparent film. Normal non-swarmed cell movement across a plate is governed by sliding motility, and produces only a slight increase in diameter from the point of inoculation (Figure 2.) Thus, bacteria that are able to swarm produces distinct patterns when grown in the laboratory that separate them from species that are unable to swarm across surfaces.
Simple Urinary Tract Infections
Urinary Tract Infections are caused by bacterial colonization of the urethra. The most prevalent agent for UTI infections are E. coli bacteria, specifically the UPEC strain, which causes over 90% of UTI infections in adults. (23) Generally, urinary tract infections are more common in women than in men, due to many physiological variations. E. coli are typically found in the gastrointestinal tract, and live in symbiosis with humans. However, upon entry into other areas of the body, they can cause many different kinds of complications. Particularly, UTIs du to E. coli are classified into three categories: Bateriuria in the urethra, Pyelonephritis in the kidney, and Cystitis in the bladder (Figure 3).
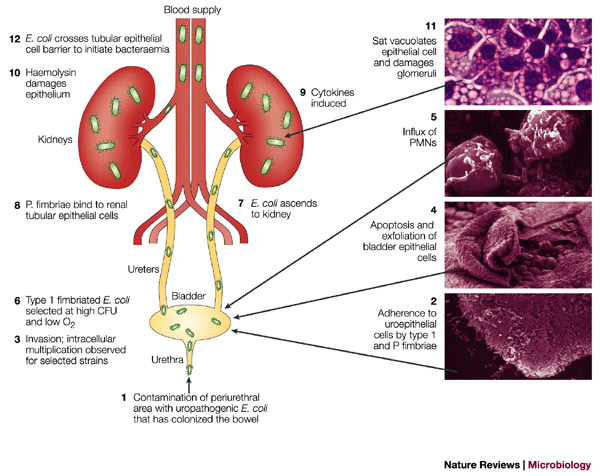
Symptoms of a urinary tract infection include the following: a strong urge to urinate, a burning sensation when urinating, pelvic discomfort, urine that appears red or cloudy, urine that is strong smelling. When UTIs infect the kidneys in pyelonephritis, the symptoms are typically more severe and include the following: upper back pain, high fever, vomiting, nausea, and chills. (24) Several risk factors may increase the likelihood of contracting a common UTI infection. Being a female, being sexually active, having urinary tract abnormalities, having a hindered immune systems, or taking birth control all increase the risk of having a UTI. (24) Urinary Tract Infections are typically diagnosed via a urine sample, in which the presence of white blood cells, red blood cells, or bacteria is screened. Sometimes, a lab culture of the urine is grown in order to determine the specific type of bacterial agent, as well as look for possible treatment options; namely, antibiotics. Sometimes, an individual who suffers from chronic UTIs will undergo a CT (computerized tomography) scan or ultrasound to have the urinary tract analyzed via images. Furthermore, a cytoscopy can be performed, which involves the insertion of a thin, long, tube into the patients urethra and bladder for further analysis. (24).
Treatment for common/simple UTIs is usually facilitated via an antibiotic regime. The most common drugs used to treat UTIs include amoxicillin, ampicillin, ciprofloxacin, levofloxacin, and Septra. (24) Symptoms usually clear within one to three days after receiving the antibiotics. In some cases, pain medicine may also be prescribed depending on the severity of symptoms. br>
Catheter Associated Urinary Tract Infections: CAUTIs
Catheter Associated Urinary Tract Infections are those that occur when bacteria enter the urinary tract via a urinary catheter. Among all urinary tract infections, it is the most rare type, especially in comparison to UTIs caused by E. coli. CAUTIs are most prevalent in hospitals or long-term health care facilities, and in patients with indwelling catheters for extended periods of time. In hospitals, CAUTIs are the most common infections that occur worldwide. The primary agents for CAUTIs are P. mirabilis, as discussed in previous sections. Virulence factors of P. mirabilis directly linked to CAUTIs will be discussed in the following section. Generally, infection begins when the bacteria enter the urethra via the catheter, and travel up the urethra via swarming activity.

Symptoms of catheter-associated urinary tract infections are similar to the symptoms experienced in simple UTIs, however are slightly more severe. According to the Journal of the Royal Society of Tropical Medicine and Hygiene, symptoms of CAUTIs typically involve the following: high or worsening fever, blood or cloudy urine, pelvic pain/discomfort, frequent urination (when catheters are removed), unexplained malaise, fatigue, or tenderness in the lower abdomen. (25) Urine samples and the production of a urine colony are used to characterize CA-UTIs in suspected patients. Treatment for complex CA-UTIs usually involves the removal of the catheter, and an antibiotic regime is usually started to kill off any remaining bacteria in the patient. In most cases, variants of CA-UTI causing bacteria have developed antibiotic resistance to many hospital antibiotics, making them much harder to treat. (25) However, several preventative measures can be undertaken to reduce the risk of Catheter induced infection. If a catheter is necessary for a hospitalized patient, intermittent catheterization may be used in place of long-term or indwelling catheters, thus reducing the risk of bacterial colonization (25). Furthermore, the recent development of antimicrobial catheters may also be used to decrease the risk of CA-UTI in hospitalized patients.
A study by Johnson et. al. (2012) compare the effectiveness of two types of Foley catheters, both of which were marketed as antimicrobial. One type was Nitrofurazone-coated, while the other was a silver alloy-coated catheter (Figure 4). Johnson et al. (2012) used an agar plate-based assay to assess the bacterial counts of four Foley catheters: one Nitrofurantoin lined catheter, and an all silicon control catheter, and one silver alloy-coated catheter and it’s latex-hydrogel control catheter. Each catheter was cut into 4 cm segments, gas sterilized, and the incubated in a tissue culture containing the usually CA-UTI biofilms. After incubation, the catheters were rinsed, and inoculum broths of the bacteria on the catheters were assessed for quantitative counts. Overall, the results showed that Nitrofurantoin lined catheters significantly decreased bacterial counts. Unfortunately, P. mirabilis , amongst other bacteria composing the Biofilm, were found to be resistant to nitrofurantoin. The study, while introducing a new and promising method of preventing CA-UTIs, also re-confrimed the incredible durability of some bacterial strains in CA-UTIs, including P. mirabilis. (26)
CAUTI Virulence Factors in Proteus Mirabilis: Pearson et. al. 2011
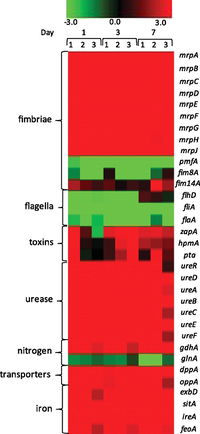
Specific virulence factors in P. mirabilis that enhance the bacteria’s pathogencity have been characterized of the course of several decades, and new virulence factors are being discovered on a yearly basis. These factors are genetically expressed when necessary during the course of the bacteria’s infection of the urinary tract increasing its virulence. In a recent study, Pearson et. al. (2011) looked into the virulence and nitrogen assimilation gene expression of P. mirabilis using a murine mouse model. The basis of this study involved characterizing and identifying the expression of virulence factors in P. mirabilis and comparing these factors to the virulence identified in E. coli (causative agent of common UTIs). In general, they identified several genes that were upregulated in the P. mirabilis UTI strain used in their mouse model: mannose resistant (Mr/P) fimbrae, iron uptake systems, protein transporters, pyruvate metabolism enzymes, TCA cycle enzymes (Figure 6). Interestingly, flagellar expression was down regulated in vivo (Figure 5). Pearson et. al. also looked specifically at genes involved in nitrogen assimilation; they found that the genes glnA and gdhA which code for the glutamine synthase and glutamate dehydrogenase, were also up regulated . The gdhA gene in particular was an unexpected find, and it is believed to help monitor the carbon-nitrogen balance in the bacteria.
To conduct their experiments, Pearson et. al. (2011) isolated P. mirabilis H14320, a strain found in long-term catheterized women. This strain was then cultured and used inoculate the urinary tract of 30 female mice. Urine samples were collected at 1, 3, and 7 days after infection. The RNA was extracted from these samples, PCR converted in cDNA, and target genes were amplified previous to microarray analysis. The measured expression from in-vivo P. mirabilis were then compared to control expression in a liquid broth, as well as the expression of in vivo E. coli. Pearson et. al. found that genes encoding MR/P fimbrae (mrpA) were the most highly regulated in vivo. Particularly, a gene named mrpI, which includes a transcriptional repressor for flagella, was up regulated in the early stages (days 1 and 3) of the infection, and then down regulated by seven. This finding suggests the importance of MR/P fibrae in the early stages of UTI infection, which is followed by the importance of flagella in the later stages of the infection. However, Contrary to previous studies, Pearson et. al. (2011) found that both flagella and PMF fimbrae were down regulated, even though they are shown to be important virulence factors in many other studies. This again may suggest their importance in the later stages of infection, since the bacteria are focused solely on the upregulation of metabolic components in the early stages.
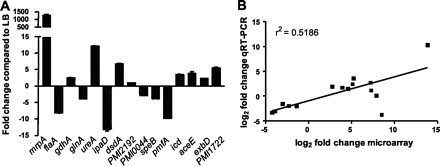
Corresponding to these metabolic changes, iron uptake genes and well as urease and glutamate dehydrogenase were found to be upregulated. Urease expression is increased in order to ensure an abundant source of ammonia in the urinary tract for the bacteria. Increased Urease expression was not found in E. coli strands, suggesting a pathogentic advantage for P. mirabilis , who are not starved for a nitrogen source when in the urinary tract. Furthermore, up regulation of nitrogen dehydrogenase (gdhA) allows for P. mirabilis to more efficiently produce urea, assimilate nitrogen for energy, and colonize the urinary tract more effectively in the long run.
Conclusion
P. mirabilis present a particular problem in CAUTIs, especially since they are highly resistant to several antibiotics in hospitals and care facilities. Their ability to engage in swarming activity proves to be advantageous, along with several other virulence factors, making them excellent candidates for CAUTIs. Current research is still being done to determine ways in the CAUTIs can be effectively treated. One future direction could possibly determine if there are antibiotics that would inhibit the expression of fimbrae, flagella, and urease that occur during swarming differentiation, and thus possibly prevent colonization of P. mirabilis.
Currently, little is understood about the swarming ability of P. mirabilis swarming motility in general. However, swarming motility is fascinating non-the-less, and continues to be a field of research for microbiologists worldwide.
References
1. K. 2013. Proteus Infections. Medscape Reference. April 22, 2013.
2. Kearns D.B. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol. 8(9): 634-644.
10. Bacheller, C. D., and J. M. Bernstein. 1997. Urinary tract infections. Med. Clin. N. Am. 81:719-730
13. Hoeniger JFM. 1965. Development of flagella by Proteus mirabilis. J. Gen. Microbiol. 40:29–42
24. Harms R.W. 2012. Urinary tract infection (UTI): symptoms. Mayo Clinic. August 29, 2012.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.