Two Modes of Secondary Metabolite Biosynthesis by Members of the Genus Streptomyces
By Samuel Kaplan
Introduction
The genus Streptomyces is the largest genus of actinobacteria, with over 500 species having been identified. Like other actinobacteria, Streptomycetes are exclusively gram-positive anaerobes, and many produce spores. “Streptomyces” translates literally to “chain fungus”, owing to the branching characteristics of its hyphae, filamentous structures that in aggregate constitute the mycelium, or the vegetative structure of the organism. Despite these morphological homologies to fungi, all members of the genus Streptomyces are decidedly bacteria (Shirling and Gottlieb, 1966).
Further, streptomycetes also exhibit some of the most diverse and complex secondary metabolic pathways of all bacteria, and as such are particularly relevant to the medical community. Many of the secondary metabolites generated by streptomycetes are known natural products, and have widespread therapeutic application as antifungal, antibacterial, anticancer, and antiparasitic drugs (Kieser, et al., 2000). Because of the wide variety within the genus Streptomyces, researchers are continually discovering and identifying novel compounds produced by streptomycetes that have potential uses in medicine. In fact, 60% of Anticancer and 70% of anti-infective antibiotics in clinical use are natural product based (Van Lanen and Shen, 2006). This fact demonstrates the obvious utility of developing a better understanding of natural-product producers such as the genus Streptomyces: with a better understanding of how these molecules are biosynthesized by microbes, researchers can begin to optimize their production and alter their structure to yield better therapeutic agents.
These secondary metabolic products have diverse structure from many standard metabolites, and as such require specialized cellular machinery. The enzymes that assemble these molecules, which occur in clusters throughout the bacterial genome, can be categorized based on mechanistic analogy. Two major routes to biosynthesis of medically-active secondary metabolites are groups of enzymes collective known as Non-ribosomal Peptide Synthetases (NRPS) and Polyketide Synthetases (PKSs) (Kieser, et al., 2000). These two highly modular synthesis pathways, along with many post-modifying enzymes such as oxygenases, oxoreductases, and glycosyltransferases, allow for the biosynthesis of a broad range of highly useful secondary metabolites (Van Lanen and Shen, 2006).
Polyketide Synthetases
Polyketide natural products are a diverse class of compounds with highly variable structure and function. Broadly speaking, polketides are molecules with at least two carbonyl groups, though most of the polyketide secondary metabolites produced by members of the genus Streptomyces have numerous other prosthetic functional groups. Such polyketide natural products display a diverse range of therapeutic properties; they are used therapeutically as antibiotics, anti-cancers, antiparasitics, and antifungals (Koehn and Carter, 2005). Because of the large size and complexity of polyketide natural products, these molecules are often produced through sequential (and often decarboxylative) Claisen condensation reactions of CoA derivatives of carboxylates (often malonyl-CoA) in a process that greatly resembles the biosynthesis of fatty acids. The sequential nature of polyketide biosynthesis allows for near-infinite additions to a growing molecule while maintaining a relatively consistent enzymatic structure. The enzymes and enzyme complexes that produce polyketides are known as Polyketide Synthetases (PKSs), and can be categorized into three morphotypes, simply called type I, II, or III, based on their catalytic domain organization. While type I PKSs are large, modular proteins that function as holoenzymes, type II PKSs are aggregates of numerous mono-functional proteins that are not bound together. This division is notable as it means type II PKSs are fully dissociable and therefore isolatable in vitro, while type I PKSs cannot be further subdivided. Type III PKSs are unique in that they do not use ACP domains (more on this later). Notably, specificity by PKSs is extremely tight, which is to say that each catalytic domain is highly selective for its specified substrate. In this sense, it is the primary structure of the protein itself which dictates the order of ketone addition, rather than any coding molecule such as DNA (Staunton and Weissman, 2001). Because each enzyme has such a specific role and these actions are carried out in sequence, PKSs can resemble enzymatic assembly lines, structures that individually make small, simple augmentations to the molecule that in aggregate generate an extremely complex finished product.
Enzymology of PKS
Despite the diverse range of molecules that they generate, PKSs have a fairly conserved enzymatic structure, consisting of seven classes of enzymes that function in three groups to finally generate a finished polyketide. At the beginning of PK synthesis, the starter acyl binds to a cysteine thiol of a ketosynthase (KS), which catalyze condensation of acyl residues. The second substrate is then loaded to the thiol residue contained in the phosphopantetheine of a modified acyl carrier protein (ACP). This post-translationally added phosphopantetheine acts as a flexible linker that can bring the developing PK chain to the requisite enzymes in order for extension to occur. The resulting condensation of acyl residues results in a β-keto ester, which remains bound to the ACP. This β-keto ester is subsequently reduced by a keto reductase (KR) and dehydrated by a dehydratase (DH) before being once more reduced by an enoyl reductase (ER) (Dyson, 2011). All of these steps, when performed in sequence, constitute a single cycle of PK extension by the PKS (Fig. 2). The growing chain passes through a number of such chain-extending modules, each of which contains at least these catalytic domains necessary for the incorporation of a ketone residue into the PK chain.
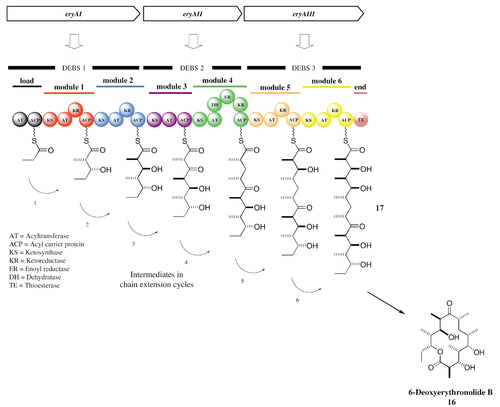
Elongation begins once the KS domain of one module receives the polyketide chain from the previous module in the synthetic pathway. The transfer of the PK occurs via trans-acylation and is catalyzed by the KS domain. The AT domain then binds the elongation group (the molecule that will become incorporated into the polyketide) to the ACP domain for loading onto the KS-bound polyketide. The ACP-bound elongation group then reacts with the KS-bound PK in a Claisen condensation to yield a new ACP-bound, elongated polyketide and a free KS domain. While the polyketide is still ACP-bound, the domains’ modification domains, such as KRs, ERs, and DHs can further alter the chemical structure of the polyketide. The process of elongation continues sequentially through all of the PKS’s domains until the TE domain hydrolyzes the finished polyketide chain from the ACP domain of the final module (Dyson, 2011).
Doxorubicin biosynthesis by S. peucetis via DXR PKS:
Doxorubicin (DXR) is a modified version of the natural product danunorubicin, a molecule produced by many strains of Streptomycetes. DXR is a potent anti-cancer drug used in chemotherapy, and functions as antibiotic by intercalating DNA and thereby preventing its transcription. The starting molecule in synthesis of DXR is propionyl-CoA, which then goes through iterative cycles of PK chain synthesis by a type II PKS in order to generate 12-deoxyalkalonic acid, which is then converted by a number of non-PKS enzymes into danunorubicin, a direct precursor to DXR; in the final step of the biosynthetic pathway, a single PKS module converts danunorubicin into DXR. While many members of the genus Streptomyces possess the requisite enzymes to danunorubicin biosynthesis, it took the work of Arcamone et al. (1969) to reveal a mutant strain, S. peucetius, which possesses the necessary enzyme to convert danunorubicin into DXR: danunorubicin-doxorubicin PKS (DXR PKS) (Arcamone et al., 1969). While researchers were able to identify this strain and most of the genes coding for DXR PKS, it took until Grimm et al. (1994) to fully characterize the genes that produce the holoenzyme. The genes were so difficult to characterize because the genes dpsG and dpsF, which encode for an ACP and a PK cyclase, respectively, are ~10 kb downstream from their usual position in type II PKSs. However, now that researchers have identified the genes responsible for DXR PKS, they have been able to use heterologous expression to more efficiently produce DXR, lowering its price per kg from $1.37 million to $1.1 million (Grimm et al., 1994).
Non-Ribosomal Peptide Synthases
A second class of secondary metabolites produced by streptomycetes is nonribosomal peptides (NRPs), which are synthesized by nonribosomal peptide synthetases (NRPSs), independent of mRNA or any other canonical translation machinery. These peptides, often less than 20 amino acids in length, have a diverse range of functions in microbial contexts. While their therapeutic use as antibiotics is the most relevant to researchers and clinicians, NRPs can also function as toxins, siderophores (iron-chelaters), or pigments. Because the biosynthesis of these peptides is not dictated by a specific mRNA sequence, NRPs often contain non-proteogenic amino acids, such as D-amino acids, and are frequently structurally modified by the addition of a number of functional prosthetic groups such as methyl, acetyl, or hydroxyl groups (Finking and Marahiel, 2004). This limitless modification of the peptide in the absence of an mRNA coding sequence affords NRPSs a high degree of freedom to produce complex, biologically and medically active natural product molecules.
The large, multi-functional NRPSs are comprised of modules, regions defined as “a catalytic portion of the NRPS that is responsible for incorporating one building block onto the growing polypeptide chain”. In bacteria, these modules are distributed over several NRPSs which are organized into an operon. Often, bacteria will use a single NRPS holoenzyme for the biosynthesis of a natural product . As a result, these proteins can reach astronomical sizes: the NRPS that synthesizes cyclosporine in T. niveum is an 11-module, 1.6 MDa behemoth. Within a module are multiple domains, each of which catalyzes a specific chemical reaction. While the enzymatic structure of each NRPS is specific to the secondary metabolite being produced, the functionalities of many domains are equivalent; these highly conserved functional sequences are known as “core-motifs” and are classified based on their functionalities (Finking and Marahiel, 2004). While these domains share much functional and structural homology with the canonical translation machinery, they lack any sequence homology with their ribosomal kin.
Enzymology NRPSs
The highly conserved “core domains” catalyze essential steps in the biosynthesis of NRPs, and can be classified by their function (Fig. 3). The NRPS’s adenylation domain (A-domain) first activates a substrate (usually an amino acid) by attaching a molecule of AMP to it. While the functions of the A-domain and aa-tRNA synthetase is fairly similar, they share little structural homology; the A-domain actually shares a high degree of sequence homology with the luciferase from Photinus pyralis. The A-domain is composed of a small C-terminal subunit and a large N-terminal subunit, and the binding pocket for the nascent polypeptide is situated where these two subunits meet. Before the substrate can bind with the A-domain, a structural element known as the P-loop, a phosphate-binding region at the entrance to the catalytic pocket, must move to open the catalytic domain. Current research suggests that the P-loop closes during catalysis in order to protect ATP from the surrounding water and begin catalysis (Strieker, et al., 2010).
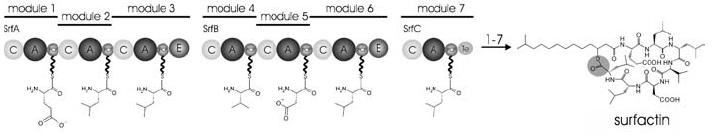
Adenylated amino acids are then attached to the 4’ pyrophosphate in a peptidyl carrier protein (PCP), whose structure is roughly analogous in function to that of a tRNA. These proteins serve to transport the activated intermediate to another PCP-AA for condensation to occur. Current research suggests that the structure of these proteins in highly pH dependent, and that their substrate binding sites can display differential specificity for various amino acids depending on environmental conditions (Finking and Marahiel, 2004). This chimeric quality of PCPs contributes to the directional biosynthesis of NRPs and also the tight regulation of their synthesis.
Condensation domains (C-domains) then catalyze nucleophilic attack of the amino group on the activated amino acid onto the acyl group of the nascent polypeptide. The peptide bond formation of these domains is directional, which is to say that the nucleophilic attack always comes from the aminoacyl residue upstream of the C-domain. The recently elucidated crystal structure of these domains reveals that they contain two key binding regions at the interface of their two major subunits: N-face, which houses the acceptor site for the upstream nucleophile and C-face, which houses the donor site for the downstream electrophile. The C-domain, too, participates in the directional synthesis of NRPs: the upstream nucleophile-binding acceptor site displays a high degree of substrate specificity, even though the downstream site, oddly enough, does not (Strieker, et al., 2010).
Termination of PCP-AA condensations is mediated by a thioesterase domain (TE-domain), which is bound the terminal PCP of the last module in the biosynthetic pathway. Termination is achieved via a two-step process that generates an acyl-O-TE-enzyme intermediate that can be attacked by either an internal, nucleophilic peptide (resulting in the formation of a cyclic peptide chain), or by a water molecule (resulting in the formation of a linear peptide chain). In vivo, cyclic molecule formations appear to be the most common form of product release, probably owing to the increased stability the cyclic structures offer over a linear structure. Owing to the diverse structure of NRPs, these enzymes have evolved to catalyze diverse enzymatic functions such as amide bond formation, lactonization, or the formation of cyclic branched structures. Because of this specialization, most TE-domains share little sequence homology (around 10%-15%) (Finking and Marahiel, 2004).
Daptomycin Biosynthesis by S. roseosporus:
Daptomycin is a cyclic lipopeptide antibiotic that is produced as a secondary metabolite by Streptomyces roseosporus via NRPSs. Is consists of ten amino acids arranged cyclically bound to a three amino acid exocyclic tail. As is characteristic of molecules produced by NRPSs, daptomycin contains two non-proetogenic amino acids: L-3-methylglutamic acid and L-kynurenine, the latter of which is found exclusively in daptomycin (Miao, et al., 2005). Initiation of biosynthesis begins with the condensation of decanoic acid with the N-terminus of tryptophan, whereupon the subsequent amino acids are added by NRPSs until termination, mediated by a TE-domain, occurs via cyclization. Three overlapping genes, dptA, dptBC, and dptD encode for the NRPS involved in daptomycin’s biosynthesis. Two upstream genes, dptE and dptF likely encode for enzymes responsible for the initiation of biosynthesis via tryptophan acetylation (Baltz et al., 2005). Once researchers cloned and sequenced the genes coding for the NRPSs responsible for daptomycin biosynthesis, researchers were able to use combinatorial biosynthesis to generate novel lipopeptide antibiotics that are derivatives of daptomycin. This ability to genetically engineer putative antibiotics from a known gene sequence affords researchers new opportunities to generate novel, highly-effective drugs through gene manipulation.
Epitoline Biosynthesis: A Fusion of Two Pathways
The pathways of PK and NRP biosynthesis can be considered highly similar: they both are highly modular processes in which sequential addition of simple, multi-carbon substrates occurs multiple times to generate a complex natural product molecule. Both pathways often generate anti-competition secondary metabolites such as antibiotics or toxins, which increase the relative fitness of the microbe relative to its neighbors (Walsh, 2004). The two processes are so similar, in fact, that emerging research has shown them to act in tandem in the biosynthesis of various molecules. The epothilones, a new class of anticancer drugs, prevent cell division by disrupting the aggregation of tubulin, the protein monomers that together form microtubules.
The drug itself was originally isolated from the soil-dwelling myxobacterium Sorangium cellulosum, and its biosynthesis involves both PKSs and NRPSs (Fig. 4). While a total synthesis of the molecule has been achieved, it is not yet economically viable, and as such Molnar et al. sought to clone and sequence the gene clusters responsible for epothilone biosynthesis.
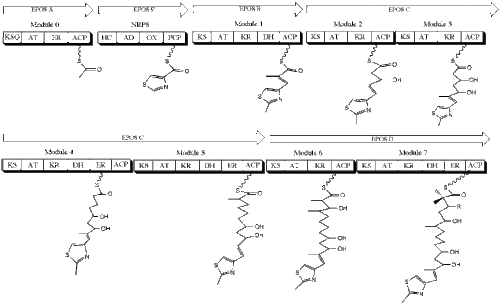
Cloning of the gene clusters involved first establishing a bacterial artificial chromosome (Bac) of the genes of analogous enzymatic structures. With computer analysis techniques such as BLASTX, researchers identified 22 putative ORFs, seven of which were identified as coding for either PKSs or NRPSs.
The canonical epothilone structure consists of a 16-membered PK macrolactone joined to a methylthiazole group by an olefinic bond. These two members are synthesized separately, with a type I PKS synthesizing the PK architecture and an NRPS incorporating the thiazole ring. The first module in the biosynthesis of epothilone is EPOS A, a PKS that contains a modified β-ketoacyl-synthase domain, an acyltransferase domain, and an enoyl reductase domain. EPOS B, an NRPS, contains an adenylation domain, an oxidase domain, a thiolation, domain, and a heterocyclization domain. These two enzymes act in tandem to biosynthesize a 2-methyl-4carboxythiazole unit, which then passes through the subsequent PKSs EPOS C, EPOS D, EPOS E, and EPOS F on the way towards becoming the finished product (Molnar, et al., 2000). With the gene clusters responsible for epothiolne identified and sequenced, researchers could now create designer epothilone derivatives via combinatorial biosynthesis.
References
Arcamone, F., G. Franceschi, and S. Penco. “Adriamycin (14-Hydroxydaunomycin), A Novel Antitumor Antibiotic.” Tetrahedron Letters, 13 (1969): 1007-1010.
Baltz, R.H. “Daptomycin: Mechanisms of Action and Resistance, and Biosynthetic Engineering.” Molecular Diversity, 13.2 (2009): 144-151
Dyson, Paul. Streptomyces: Molecular Biology and Biotechnology. (2011). Norwich, England: Horizon Scientific Press.
Finking, R. and M.A. Marahiel. “Biosynthesis of Nonribosomal Peptides.” Annual Review of Microbiology, 58 (2004): 453-488.
Grimm, A., K. Madduri, A. Ali, and C.R. Hutchinson. “Characterization of the Streptomyces peucetius ATCC 29050 genes encoding doxorubicin polyketide synthase.” Gene, 151.1-2 (1994): 1-10.
Kieser, T., M.J. Bibb, M.J. Buttner, K.F. Chater, and D.A. Hopwood. Practical Streptomyces Genetics. (2009). Norwich, England: John Innes Foundation.
Koehn, F.E. and G.T. Carter. “The Evolving Role of Natural Products in Drug Discovery.” Nature Reviews Drug Discovery, 4 (2005): 206-220.
Miao, V., M. F. Coeffet-LeGal, P. Brian, R. Brost, J. Penn, A. Whiting, S. Martin, R. Ford, I. Parr, M. Bouchard, C.J. Silva, S.K. Wrigley, and R.H. Baltz. “Daptomycin biosynthesis in Streptomyces roseosporus: cloning and analysis of the gene cluster and revision of peptide stereochemistry.” Microbiology, 151.5 (2005): 1507-1523.
Molnar, I., T. Schupp, M. Ono, R.E. Zirkle, M. Milnamow, B. Nowak-Thompson, N. Engel, C. Toupet, A. Stratmann, D.D. Cyr, J. Gorlach, J.M. Mayo, A. Hu, S. Goff, J. Schmid, and J.M. Ligon. “The biosynthetic gene cluster for the microtubule-stabilizing agents epothilones A and B from Sorangium cellulosum So ce90.” Chemistry and Biology, 7.2 (2000): 97-109.
Staunton, J. and K.A. Weissman. “Polyketide Biosynthesis: a Millenium Review.” Natural Product Reports, 18 (2001): 380-416.
Strieker, M., A. Tanovic, and M.A. Marahiel. “Nonribosomal Peptide Synthetases: Structures and Dynamics.” Current Opinions in Structural Biology, 20.2 (2010): 234-40.
Shirling, E.B. and D. Gottlieb. “Methods for Characterization of Streptomyces Species.” International Journal of Systematic Bacteriology, 16.3 (1966): 313-340.
Van Lanen, S.G. and B. Shen. “Microbial Genomics for the Improvement of Natural Product Discovery.” Ecology and Industrial Microbiology, 9.3 (2006): 252-260.
Walsh, Christopher T. “Polyketide and Nonribosomal Peptide Antibiotics: Modularity and Versatility.” Science, 303.5665 (2004): 1805-1810.
Edited by student of Joan Slonczewski for BIOL 238 Microbiology, 2009, Kenyon College.
