Sargasso Sea


Introduction
Deep within the Atlantic Ocean, near the Bermuda Triangle, lies a sea shrouded with mystery named the Sargasso Sea. Uncannily calm, the Sargasso Sea was believed to be the demise of sailors. With little wind, ships lay stagnant for days in this “sea of lost ships,” leading to historical accounts of crews going into the sea, but never coming out. Eventually these accounts were mutated into urban legends of whole ships disappearing or broken down vessels roaming this sea manned by skeleton ghost crews and all (The Sargasso Sea). Realistically speaking though, the Sargasso Sea was believed to be first found by Christopher Columbus and his crew. They named this sea after the seaweed dominating the sea’s surface named Sargassum. They named the sea “Sargaco”, which means grape because Sargassum looks like grapes (Gladnick, 2008).
Description of Niche
Where is it located?
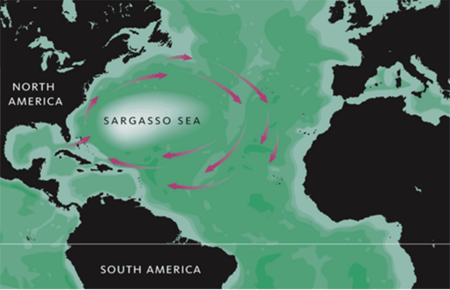
More precisely, the Sargasso Sea is located in the middle of North Atlantic Ocean bounding the Great Antilles on the south, the Gulf stream on the west, and Bermuda on the north (Teal & Teal, 1975). These currents move around the Sargasso sea in a clockwise orientation. Having a latitude between 20N and 35 N and a longitude between 30W and 70W, the Sargasso Sea is comparable to the United states in size (Teal & Teal, 1975). Both this great size and the location of the Sargasso Sea have great implications on its physical conditions.
Physical Conditions?
Because it is not bounded by coastline and is surrounded by strong currents, the Sargasso Sea is considered an isolated, oligotrophic (little to sustain life) area. The surrounding currents provide a strong physical boundary separating the Sargasso sea from the more nutrient-rich waters of the North America shelf. The northern region contains warm water known as eighteen-degree water that moves outwards along the surface of the sea, allowing it to maintain that temperature year round whereas coastal waters with the same latitudes freeze in the winter. With increasing depth, water temperature declines and pressure builds up restricting much life (Teal & Teal, 1975). Also, the water in the Sargasso Sea is said to be salty and warm, maintaining a salinity around 36% and euphotic zone temperature up to 22°C. For these reasons, even though there are several species of plankton and massive amounts of seaweed floating on the water surface, the Sargasso Sea is still not nutritious enough to attract large communities of fishes. These factors of low wind, low nutrients, and high salinity help to justify why the Sargasso Sea is considered a desert of oceans with little sign of life.
As mentioned before, little nutrients are detected in the Sargasso Sea. One conformation of this belief is Phosphorous amounts. Studies conducted on the Sargasso sea suggest a severe phosphorus depletion and that the cause of this phenomenon results from the large difference between the DNN (dissolved nitrate plus nitrite) to DIP (dissolved inorganic phosphate) ratio. This large discrepancy causes P to be depleted long before N is consumed by algal growth after the nutrient injection from the aphotic zone (Wu et al., 2000).
Another type of nutrient found in the Sargasso Sea are slicks, natural fat and oil buildup. Not only do these nutrients contribute to the sea’s overall calm glasslike surface, bacteria can collect and grow on these leveled surfaces. They contain abundant useable organic materials; and, once the bacteria dies, the bacteria yields even more oil to these slicks. Unfortunately, one slick area does not last long and tends to fade away gradually due to various chemical and bacterial activities (Teal & Teal, 1975).
How Research Began
To understand what is known about the Sargasso Sea, one needs to understand why we wanted to know more about it in the first place. Although most scientists thought that this expanse of sea contained no sustainable life, observations showed that oxygen and other elements were being consumed at a higher rate than theories and models could account for. This led scientists to think there must be some nutrient source fueling the blooms of phytoplankton in the Sargasso Sea ( Carlowicz, 2006; LiveScience). This discovery, in turn, led to the discovery of eddies.
Eddies and Their Influence on the Sargasso Sea
Even with such a hostile environment, there exists vast phytoplankton bloom and higher oxygen consumption rate than predicted in the surface euphotic area. The vast amount of organisms in the Sargasso Sea documented in Venter et. al is shocking in comparison to the lack of “nitrate, phosphate, trace metals, and other nutrients” present (Carlowicz, 2006). This suggests that there might be internal water mixing that could continually pump up rich nutrients from deeper water layers. This swirling water system is called eddies (LiveScience). Eddies are episodic underwater current systems that forcefully bring nutrients up from the ocean floor. They form from “differences in ocean temperature and salinity that give water different densities. Like oil and water, water masses of different densities tend to keep separate, rather than mix” (Carlowicz, 2006). This brings necessary food to the phytoplankton and other microbes. Specifically, during the winter, the incoming cold water forms a “subtropical mode,” that allows phytoplankton to multiply due to the increase of nutrients. What causes eddies to spin and therefore mix during the summertime is the Earth’s natural rotation, also known as the Coriolis force (Carlowicz, 2006). This results in an increase in other organisms such as zooplankton that ultimately feed the entire environmental niche (Venter et al., 2004; LiveScience). This also fuels sargassum growth and without this, organisms living within the sargassum would not be able to survive (Sargasso Sea Without a Coastline). Therefore eddies, often called “the oasis of the ocean”, can be essential to many microbes living in the Sargasso Sea (LiveScience).
Genomic Revolution
In order to identify and quantify the microbial life in the Earth’s oceans, a metagenomics research project called the Global Ocean Sampling (GOS) survey was undertaken by J. Craig Venter et al. Thinking that the initial project would be an easy task in the Sargasso Sea because of the lack of nutrients; they soon found it to be daunting task (Ruder, 2004).
As mentioned Venter is classifying species by their DNA and genes instead of appearance in order to understand their evolutionary existence in their respective environments. For the Sargasso Sea research, Venter accomplished this utilizing the “shotgun sequencing method to map the DNA of the microbes” which is the same methodology Venter used to map the Human genome (Ruder, 2004). This recent research by Craig Venter and his team are noted for their innovative microbial discovery by again means of genomic shotgun sequencing by exploring the Sargasso Sea and documenting unexpected amounts of various microbes and organisms. The goal was to test the usefulness of genomic shotgun sequencing technique on an environment’s species and microbes such as the Sargasso Sea’s (Venter et al, 2004).
Based on their findings, it was shown that there is a diverse amount of prokaryotic life in the Sargasso Sea then previously thought leading researchers to the idea that the diversity and abundance of ocean life is larger than once thought (Wikipedia.com, Ruder, 2004). Specifically, Venter et al. “found 1800 species of microbes, including 150 new species of bacteria, and over 1.2 million new genes (Ruder, 2004).”
The hope is that with further exploration and the discovery of various microbes and bacteria able to synthesize energy by means of carbon cycling and generating hydrogen through the use of light sources, the Venter Institute can understand more about the vast microbe diversity and their influence in their environments as well as for uncovering new methods of energy formations as well as alternative energy productions (Frazier, 2005).
Who lives there?
Several species of the brown algae belonging to the genus Sargassum are found in the world’s temperate and tropical oceans. The main species found on the Sargasso are Sargassum natans and Sargassum fluitans – both of which are completely adapted to living in the open ocean (Coston-Clements et al., 1991).
Cyanobacteria such as Dichoothrix and Oscillatoria are primarily involved in the N2 fixation within the Sargassum community. (Coston-Clements et al., 1991).
The fact that the rate of water evaporation in the Sargasso is faster at its center than at the surrounding waters generates an inward current that aggregates the countless tons of kelp in the middle of the Sargasso sea. Sargasso – or gulf weed – with its complex branching systems, allow tiny animals to hide from predators and serve as a nursery for dozens of fish species. Their blades are the only solid surface for hundreds of miles and they are the only place where sedentary animals can become attached. This community functions as a filter that sieves out nutrients and particles from the waters creating a complex ecosystem where all animals are intertwined. However, not all life forms in the Sargasso are minuscule, sea turtles, dolphins and even humpback whales find the Sargasso to be a haven – it was there that their music was first recorded (Genthe, 1998).
The sea’s food chain in which all existing animals rely on for survival, starts with the surface plants acquiring energy and sunlight. The plant tissues grow, containing various synthesized chemicals that animals can consume and pass along as they are eaten themselves. However, the process of vertical migration is very gradual and slow in the sea. Vertical migration is the downward transport of assorted animal species who have consumed other animals or phytoplankton and will be eaten themselves by the lower-dwelling animals; this process occurs in order to obtain food and gain energy. This food chain allows the sea’s animals to coexist and depend upon one another for food and energy gain (Teal & Teal, 1975).
Which microbes are present?
Prokarya
One of the most interesting microbes comes from the Roseobacter clade, of α-proteobacteria. Silicibacter pomeroyi is one of the most numerous oligotrophs that inhabit the Sargasso Sea. Having evolved in an area of the world with low nutrients, it has developed a unique physiology compared to other marine oligotrophs. Some of these include; sulfonate degradation, oxidation of lignin-related compounds, and the use of hydroxylamine oxidoreductase to generate nitrite. These characteristics allow Silicibacter to cope with the under nourishing habitat. The microbe also contains genes that enable a symbiotic relationship with plankton, uptake algal produced compounds, fast growth and cell density dependent regulation (Moran et al., 2004).
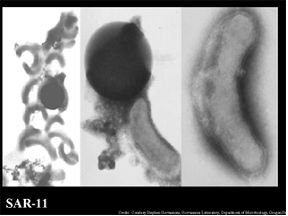
SAR Family of Bacteria
One of the documented most abundant microbes of the marine world, as well as the Sargasso Sea, belongs to the clade of SAR11 α – proteobacteria. The name “Sar” was given to this family of bacteria due to their discovery in the Sargasso Sea in 1990. One species that is of most interest to researchers is that of Pelagibacter ubique. P. ubique covers 30% of the surface in the Sargasso. During the summer months the population of this bacterium can cover up to 50% of the ocean’s surface weighing more than the weight of all of the fish in the all of the collective oceans (Giovanni et al., 2005). What make these bacteria an interest of research, is its ability to thrive in an environment low in nutrients and resources. This bacterium can surprisingly replicate efficiently in a low nutrient environment and is one of the smallest self-replicating cells found. Evolutionary genome reduction has been observed in this microbe. This is consistent with the hypothesis of “genome streamlining driven by selection acting on a very large population which resides in a very low nutrient habitat”. The belief is that the bacterium’s genome is being reduced so not to expend energy on replicating DNA with no adaptive value. This saves the the organism from performing unnecessary metabolic tasks (Giovanni et al., 2005).
SAR86 is part of the Proteorhodopsins (PRs) family-related species. PRs are predicted to have important roles in supplying light energy for microbial metabolism. They are divided into two groups depending on two distinct absorption spectrums of light intensity: Green-absorbing (GPRs) and Blue-absorbing (BPRs). In the Sargasso Sea, the dominant light energy at surface water (until 40m depth) is in the blue range with maximal intensity near 440m. Therefore, only BPRs are discovered regardless of seasonal changes and depth of the surface water column, an indication of different light intensity. This is a unique feature of the Sargasso Sea that is different from other ocean areas, such as Mediterranean Sea, where you will find GPRs dominant in the upper most water layer because of the presence of green pigments, then followed by BPRs in the next water layers (Sabehi et al., 2007). This adaptation to spectrum changes in the environment is called spectral tuning, which requires a single amino-acid change at position 105: leucine for green and glutamate for blue (Man et al., 2003).
Cyanobacteria
Though known as the smallest photosynthetic organism ("Prochlorococcus marinus NATL2A"), Prochlorococcus is still one of the major phototrophs in the ocean and greatly impacts the carbon cycle. (Partensky, 1999;Campbell, 1994). It is a small photosynthetic marine cyanobacterium found up to 200 meters below the sea surface throughout the euphotic zone (Partensky, 1999;Campbell, 1994). Though a cyanobacteria, Prochlorococcus has a unique pigment composition. It contains divinyl derivatives of chlorophyll a and b, α -carotene, zeaxanthin (Chisholm et al., 1992; Goericke, 1992) and phycoerythrin (Hess et al., 2001). Growth of Prochlorococcus is best when surface waters are devoid of major nutrients and worst during winter when Synechococcus and other eukaryotic organisms have their plankton bloom (DuRand et al., 2001; Zubkov, 2000; Olsen,1990). Prochlorococcus have different ratios of chlorophyll pigments based on where they live. High-light adapted Prochlorococci live in the upper 100 meters which are well lit. Low-light adapted Procholorococci are found mainly in deep euphotic zone of depth 80-200 meters, which is a less bright environment but nutrient rich (Dufresne et al., 2003). Low-light adapted Prochlorococci are generally found to be able to take up NO3- (Wyman, 1985).
Another major contributor to the Sargasso ecosystem and closely related to Prochlorococcus is the cyanobacteria Synechococcus. Though less abundant than Prochlorococcus, Synechococcus has a larger cell size and broader lateral distribution (Herdman et al., 2001) therefore giving it the ability to compete metabolically with its more numerous counterpart, Prochlorococcus. Synechococcus is restricted to living in the upper 100 meters under the sea surface (Bertilson et al., 2003; Durand et al., 2001). Unlike Prochlorococcus, Synechococcus contains a pigment composition typical of cyanobacteria consisting of chlorophyll a and phycobilins. It is known to be good at obtaining trace metals and major nutrients even in oligotrophic environments such as the Sargasso Sea (Casey et al., 2007).
Luminous Bacteria
Vibrio fischeri and Lucibacterium harveyi consist of 90% of the luminous bacteria isolated from Sargasso Sea. Photobacterium leiognathi and Photobacterium phosphoreum constitute the rest of the population. Only Vibrio fischeri and Lucibacterium harveyi are found in the upper water layer, at depths of 160m to 320m. The average temperature here,at 20C, is where thermocline occurs. No luminous bacteria are found in surface microlayer. Luminous bacteria are small rod shape, gram negative bacteria. They could be free-living planktons, parasites or they can symbiont with fishes. Their major role is the production of bioluminescence which serves as specific functions for different species. It’s said that free-living life style is less likely to occur in the Sargasso sea since it’s an oligotrophic area, therefore the majority of them have a symbiotic relationship with small sea animals that live within the Sargassum (Orandorff & Colwell, 1980).
Burkholderia is a rod-shaped, gram-negative proteobacteria. They are generally obligately aerobic and microorganisms of this type found in the Sargasso Sea are of a species similar to its terrestrial counterpart. Burkholderia bacteria are commonly found in the soil and in groundwater worldwide and it was unusual when scientists found it in the Sargasso Sea. The fact this new species of Burkholderia contains similar DNA and genes to its land species suggest genomic transfer. Terrestrial Burkholderia bacteria are known to be “good biodegraders of polychlorinated biphenyls (PCBs).” Its genome is large, “the largest of the soil bacteria known, may contribute to its ecological success (Burkholderia).” Because this and related PCB-degrading bacteria have such important ecological and commercial potential for bioremediation, sequencing the Burkholderia genome will play a major role in gaining insight on the mechanisms for its environmental versatility.
Shewanella
The Shewanella bacterium, initially found in the freshwaters of the Oneida Lake, thrives in high nutrient aquatic conditions and is capable of cycling organic carbon owing to its numerous respiratory abilities (Yang et al, 2006). Recently discovered in the Sargasso Sea are bacterial strains with genomic sequences directly associated to that of the Shewanella oneidensis bacterium. The existence of S. oneidensis deviates from expectations considering these bacteria typically exist in waters consisting of large amounts of nutrient sources which the Sargasso Sea is thought to lack much of. (Venter et al, 2004). Recent studies reveal that this bacterium is very promising to the beneficial process of bioremediation. S. oneidensis is capable of anaerobic respiration and possesses surface proteins which carries out electron transport as well as minimizes toxic amounts of substances containing metals, as a result preventing its increasing occurrence in the environment (Tang et al, 2006).
Eukarya
Diatoms and coccolithophores are among the best known brown algae found on the world’s oceans. Diatoms found in the Sargasso are curiously extremely small and have sizes in the range of small prokaryotes. Coccolithophores are also found throughout the Sargasso and were named after the tiny calcium structures that compose their outer skeleton - the coccoliths (Stingl et al., 2007).
Viruses
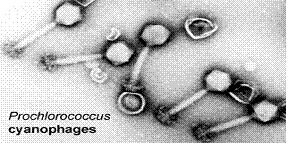
Viruses are known to be the most numerous biological entities on the world’s oceans and Cyanophages has been proved to be the dominant kind on the Sargasso’s waters. Cyanophages – as their name implies – kill cyanobacteria, thus they are actively involved in the control of cyanobacteria numbers as well as their evolution through genome insertion. Through lysogenic cycles, cyanophages are responsible for many alterations found in different strains of the same microbe species – they are responsible for moving genes across different cells which speeds up evolution (Angly et al., 2006).
Non-Microbes: Plants and Animals
It is safe enough to assume that all living organisms living on the surface of the Sargasso sea are directly or indirectly dependent on the organisms mostly associated with it - Sargassum weeds. Sargassum is known to harbor epiphytes (micro and macro), diverse fungi, over a hundred invertebrate species, dozens of fishes, and about four sea turtles species - one of which is the famous green turtle (Chelonia mydas) (Coston-Clements et al., 1991).
Among the diverse organisms living within the Sargasso Sea include sea turtles and eels. After a sea turtle hatches, it goes into the sea and migrates to an area with favorable conditions. The turtles generally spend 2-4 years here then migrate back to the North American coast (The Sargasso Sea). During their first year of life, turtles associate themselves with Sargassum rafts, mats of brown algae, about 7 million tons of which can be found in the Sargasso Sea (Milton et al., 2003). Though different species of sea turtles aquire different diets as they grow older, Sargassum provides food and shelter for the sea turtles which is desperately needed during their early years (Milton et al., 2003). To get to the Sargasso Sea, turtles use earth’s magnetic field as guidance. This is extremely important because although the Sargasso Sea and the rest of the North Atlantic gyre have a favorable environment for the sea turtles to grow in, going north of the gyre to lower temperatures could be detrimental to the turtle’s survival (Lohmann et al., 2001).
Unlike sea turtles, who migrate to the Sargasso Sea as juveniles, for eels the Sargasso Sea is a place to spawn. Two major species of eels are found here, the European eels Anguilla anguilla and the American eels Anguilla rostrata (Rosalind, 2006). Eels have a variety of diets based on age and species. For instance, American eels will eat large invertebrates as adults and phytoplankton as babies. After birth, the eels head toward Europe or North America and come back later in life to lay their own eggs (McCord). Sadly, there seems to be a decline in eel recruitment which scientists believe to be linked to climate changes in the Sargasso Sea (Changes in Ocean Conditions in Sargasso Sea).
Interactions: Competition and Symbiosis
Prochlorococcus cyanophages are known to infect Synechococcus and Prochlorococcus of both high-light and low-light adaptations. Different cyanophages can bind to different types of organisms with different specificities. Some cyanophages are host specific while others can cross infect other species such as Synechococcus (Sullivan, 2003). Podoviridae is host specific for high-light- adapted Prochlorococcus while Myoviridae affects a broad range of hosts including low-light-adapted Prochlorococcus and Synechococcus. (Sullivan, 2003) Concentration of cyanobacteria is environment specific, decreasing when going from coastal to open ocean ecosystems such as the Sargasso Sea. Scientists aren’t sure why this occurs, but one hypothesis is that cyanophages decrease production because of a decrease in host cell growth. This could be due to a decrease in nutrient availability (Sullivan, 2003).
Silicibacter has a unique evolutionary symbiosis with eukaryotic organisms. It contains a Type IV secretion system in the bacterial genome. The genes encode for DNA export to eukaryotic cells (Dolowy et al, 2005). For example Agrobacterium tumefaciens is plant pathogen. It is able to insert it’s DNA into a plant cell. Unfortunalely, this is bad for the plants, but the exhange between marine organisms is beneficial to the eukaryote (Christie et. al, 2005). This gives widespread ability of the taxon to transfer DNA or proteins to other plankton. It is thought that this system has evolved due to bacterial-dinoflagellate interactions (Worden, et al 2006).
Microbial Metabolic Effect on the Sargasso Sea
Bacterioplankton: Carbon Fixation
Not only do microbes within the Sargasso Sea affect the environment they live in, the environment reciprocally affects microbes in a similar manner. An example of such are bacterioplankton. Baterioplankton are important in regulating the amount of DOC, dissolved organic carbon, within the Sargasso Sea. In fact, the uptake through bacterioplankton of DOC is believed to be the primary consumption method of DOC (Carlson et al., 2002). Many factors play into how DOC is consumed including nutrient uptake and bacterial community structure. A common theory, the malfunctioning microbial loop theory, follows the belief that adding inorganic nitrogen or phosphorous will stimulate the growth of bacteria such as bacterioplanktons (Thingstad et al., 1995, 1997). This in turn would affect the amount of DOC exported and organic carbon stored in the Sargasso Sea. Cotner et al. (1997) and Rivkin and Anderson (1997) have done experiments showing that limitations in Phosphorous is a rate limiting factor in bacterioplankton growth. Also, the structure of the community these bacterioplankton create affects the types of nutrients they absorb (Giovannoni et al. 1996, Karner et al. 2001). This makes community structure important in DOC consumption. Other scientists believe that there is an aspect of the DOC substrate which prevents the bacterioplankton from absorbing it quickly therefore affecting the amount of DOC in the environment (Carlson et al., 2002).
Prochlorococcus & Synechococcus: Carbon and Nitrogen Cycles
Photosynthetic organisms near the surface of the ocean create DOC, dissolved organic carbon, and other organic material. Phytoplankton, such as diatoms, dinoflagellates, and cyanobacteria do much of this conversion (Carbon Cycling and Sequestration continued, 2005). Dominant among these photosynthetic organisms are cyanobacteria Synechococcus and Prochlorococcus (Carbon Cycling and Sequestration continued, 2005). Variations in phytoplankton amounts can affect the ability of the ocean to take up atmospheric carbon. Much like bacterioplankton and other heterotrophs, phytoplankton are believed to be affected by similar nutrients supplements such as Phosphorous. Prochlorococcus is known to grow well on NH4+ and only low-light types are able to grow on NO2-. It has been shown some Prochlorococcus also utilize urea along with NH4+ (Moore, 1995;Palinkska, 2000;Rippka, 2000). Currently research is being done to show that there are different ecotypes of Prochlorococcus based on the depth that the organism lives in, leading to differences in nitrogen uptake (Casey, 2007). Ecotypes are organisms from the same species that are genetically different due environmental adaptation.
Synechococcus, depending on its species, is able to utilize “NH4+, NO3-, NO2-, urea, and amino acids” as its carbon source (Goericke, 1992). When there is no nitrogen, Synechococcus will degrade phycoerythrin, a light harvesting pigment housed in it. Phycoerythrin can then be used as a replacement nitrogen source (Moore, 2002).
Trichodesmium: Nitrogen Cycle
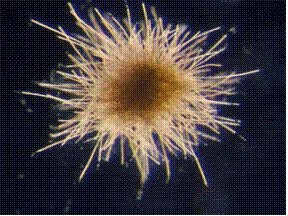
A type of cyanobacteria known to fix N2 in the Sargasso Sea is the Trichodesmium bacterium. It is found in all tropical and subtropical oceans of the world and is known to form large colonies of filaments up to 200 cells. There have been reports of blooms as large as 100,000 km2 caused by Trichodesmium thiebautii (Sudek et al., 2006). It is known to scientists due to its high contribution of N2 fixation in the tropical oceans. A hot topic amongst scientists that study this microbe, regard how their fixation rate is constrained or boosted. New findings and discoveries have highlighted the fact that phosphorus is crucial for N2 fixation and that high-affinity mechanisms that capture phosphorus in the surrounding waters are key elements that allow them to thrive in the low-phosphorus waters of the Sargasso (Haley & Dyhrman, 2006). They also have a high requirement for iron (up to 10 times higher than other autotrophic organisms) which is necessary for the assemblage of the nitrogenase enzyme. Where does this iron source arise from? Scientists have found that enough iron is transported to the Sargasso through winds that carry sand and dust from the Sahara (Langlois et al., 2008). This cyanobacterium, which is believed to fix an enormous portion of the world’s N2 – about 40% – has attracted not only the attention of many scientists, but also of the general public due to several reported coastal algal blooms over the past years which have affected both humans and animals directly (Tamandare fever) and indirectly by stimulating blooms of other harmful bacteria (Sudek et al., 2006).
Pelagibacter ubique: Carbon, Sulfur, and Nitrogen Cycles
Having an extremely simple metabolism and small genome size, Pelagibacter ubique mimics marine unicellular cyanobacteria. Due to low nutrients in the surrounding environment, P.ubique has developed metabolic pathways for 20 amino acids and almost all of the known cofactors (Malmstrom, et. al, 2004). One of the main trophic cycles it is involved with is the recycling of organic carbon. This restoration of organic carbon back into the environment provides carbon used by photosynthetic algae. This algae produces much of the world’s oxygen. Like their other α – proteobacterial countertypes, P.ubique has the ability to break-down and use the component compounds of DMSP. The break-down process is extremely similar to that of Silicibacter pomeroyi, except the latter is able to break down DMSP more efficiently for usage by other microbes. A great deal of the DMSP is used for respiration in P.ubique and limits the amount that is available for the other organisms. Amino acid usage is by far more commonly used in P.ubique than any other organism (Malmstrom, et al., 2004). Dissolved amino acids provide 5-55% of the entire bacterial carbon and nitrogen demand. Since, a majority of the DMSP is used for respiration this bacterium counts on amino acids to supply it with energy. Between the DMSP and all of the amino acids, almost all of its carbon, nitrogen, and sulfur needs are met. Also of interest, researchers can use P.ubique to study the global sulfur cycle. Emissions of DMS are thought to be responsible for increasing the Earth’s cloud cover and its ability to reflect the sun’s light (Malmstrom, et al., 2004).
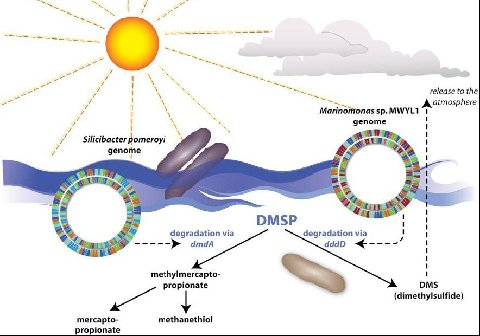
Silicibacter pomeroyi: Carbon and Sulfur Cycles
One important effect Silicibacter has on the environment is the breakdown of the compound Dimethylsulfonioproprionate (DMSP). There are two possible fates of DMSP. The first pathway breaks DMSP down into Dimethylsulfide (DMS). DMS is an extremely volatile compound readily transported from the sea to the atmosphere. This pathway is the result of degradation by Marinomonas sp. of bacteria. The second, more desirable pathway is the route in which Silicibacter uses the products of the reaction. DMSP is disassembled into less volatile components, (see figure) that are able to be used by the surrounding bacteria. This non-DMS type of synthesis allows sulfur and carbon to be provided to the marine microbial food web. This is an important part of the sulfur cycle and aids in the biogeochemical processes in the environment. Either of these routes, which end up dominating the surface of the marine water, leads to major implications in regards to the regulation of global temperature. When DMS is exchanged across the boundary (from ocean to atmosphere), it drastically affects the global temperature and cloud formation (Todd et al. 2007).
Silicibacter pomeroyi has the ability to oxidize CO to CO2 by way of CO dehydrogenase making it into organic carbon. There is an absence of ribulose bisphosphate carboxylase implicating that the bacteria gains its energy but not its carbon, from carboxidotrophy. Silicibacter-like lithoheterotrophs that share the Sargasso Sea which creates a carbon monoxide basin near the surface of the ocean. The relative number of Silicibacter is not enough bacteria to fix the entire amount CO, so it is assimilated into the atmosphere. The caveat this brings is the protection of partial pressure of greenhouse gas in the ocean (Moran, et al. 2004).
Current Research
Genetic Analysis
A new way to explore microbial ecology and to discover new species from environmental sample collections
A study by Hooper et al. has recently critically analyzed the Baas-Becking hypothesis which states that “everything is everywhere: but the environment selects” (O’Malley 2007) in relation to the presence of microbes in specific environments. This study on comparative metagenomics takes two very different environmental sequencing projects, the first from Minnesota farm soil and the second from the Sargasso Sea microbes by Venter et al., and compares the sequencing projects to find “molecular evidence of a transfer of microbes over distant environments” (Hooper, 2008). The theory behind this is that genetic material can be anywhere, yet this does not necessarily mean it will survive in a new environment but can still contribute its genome. To determine this, they measured three distinct characteristics, guanine/cytosine (GC) content, oligomer frequency patterns (OFPs), and lastly protein similarity between translated open reading frames in both data sets (Hooper et al., 2008). The research concluded with its findings that microbe transfer is a possible mechanism as similar species from land and sea have been found to inhabit both niches while maintaining similar genetic make-up.
Later on, by applying shot-gun sequencing and phylogenetic comparison on uncultured microbes collected from ocean samples, the scientists discovered several previously unknown species that don’t grow in laboratory condition. For example, in 2007, Not et al., showed the discovery of Picoplanktonic protists. By 18s rRNA analysis of different water columns in Sargasso Sea, species in Kingdom Chromalveolata and Rhizaria are found to be the prodominent protista. Among Chromalveolata, Stramenopiles, which exist only in surface euphotic area, and Alveolata, which are discovered from surface to deep sea area, are dominant species. Many of which contain choloroplast; thus living as autotrophs carrying out photosynthesis. Radiolaria is the major species found in Kingdom Rhizaria, they live mainly from 500m thermocline to 3000m deep sea and are heterotrophic zooplanktons. They may be important to ocean biogeochemical cycling and carbon transport (Ghedin & Claverie, 2005). (please refer to microbewiki for details in these species)
Not et al., utilized the similar method, genome sequence analysis and phylogenetic comparison analysis to unveil the existence of Mimivirus relatives in Sargasso Sea, a DNA virus of 0.1 to 0.8 microns in size. They are evolutionarily closer to Mimivirus and exist in large abundance in the collected water samples. The discovery is published in 2005 and further research is needed (Not et al., 2007).
Metabolic and Proteomic Analysis
SAR11 Bacteria and the State of the Ocean
Dr. Stephen Giovanni and his cohorts have done extensive research on the SAR clade of bacteria. Of special interest to them is the Pelagibacter ubique organism. Currently they are trying to use P.ubique for a few different research interests. First, they are trying to predict the different organic carbon sources used by P.ubique. They are doing this through a method of metabolic reconstruction. Second, they are using mass spectrometry to understand P.ubique’s regulatory responses in regards to environmental factors. By doing this, it will give the researchers insight to the proteomic state of the organism. With this they can then use P.ubique as a proxy to report the biological state of the specific system they are in. Last, a long-term goal is to be able to model the metabolic processes of P.ubique. It is an ideal candidate because it is one of the smallest and simplest known cells. With this information, it could be possible to integrate and optimize metabolic processes to be more efficient during low nutrient periods (Monterey Bay Aquarium Research Institute).
Unique Geological Feature: Floating Plastic
Frequently discovered throughout the western regions of the Sargasso Sea are unusual amounts of broken plastic fragments afloat on the water surfaces, roughly 3500 per square kilometer. Since these plastic fragments have become apart of the sea, various groups of hydroids and diatoms are often found on them. There are concerns regarding the ever increasing plastic production along with its improper disposal methods, which can disrupt ocean life and harm animals that may ingest plasticizers such as polychlorinated biphenyls (PCB’s) (Carpenter & Smith, 1972). Researchers inspecting the Sargasso discovered on average, about 8,000 to 10,000 of floating plastic pieces per square mile (Murphy, 1986). In addition, the Sargasso Sea consists of many plastic fragments due to its slow circulation (Greenpeace International) and its northern region includes one main, circularly moving gyre in which waste and plastic pieces flows to and accumulates (Watson, 2006).
Unique Biological Feature: Antioxidant and Antibacterial Activity
It has been found that seaweed of the genus Sargassum yields a methanol extract with potent antioxidant and antimicrobial properties. When compared with Ampicillin, it shows a more pronounced bactericidal effect on gram-positive and gram-negative bacteria. Scientists were able to evaluate these properties against three different species of bacteria, namely, Staphylococcus aureus, Bacillus subtilis, and Escherichia coli, the last being among the most common cause of food poisoning. For this reason, it has been suggested that the methanol extract of Sargassum sp. should be utilized as a cheap and plentiful natural source of food with the benefits of having antimicrobial and antioxidant properties. Nonetheless, more studies are required due the fact that the precise mechanism of how it works against bacteria is not well understood as well as possible allergic reactions that may arise (Patra et al., 2008).
Conclusion
Genomics allows researchers to find evidence of genes that are the basis of “biogeochemical activities of ocean-dwelling microbes” (Moran, 2007). Realistically taking on the task to genotype marine microorganisms has been made possible through “whole-genome shotgun sequencing” where target genes can remain unknown without impairing the search for novel genes (Venter et al, 2004). This has shed light on many new and unknown micro-organisms as only a fraction of the microorganisms found in its primary environment cannot be grown successfully in pure culture (Moran, 2007).”
To sum it all up, the culmination of data that has arisen in the past decade after the initial study by Venter et al., has shed light into the oceans unexplored biodiversity. The study of the Sargasso Sea has brought more questions to the table that researchers are currently tackling. Questions such as: “What else in the ocean don’t we know about? Are there many more genes in marine organisms than we ever imagined? Have we missed the major players in the ocean’s energy cycles?” have scientists baffled at the possibilities (Ruder et al., 2008).
References
Angly, Florent E., Felts, Ben, Breitbart, Mya, Salamon, Peter, Edwards, Robert, A.Carlson, Craig, Chan, Amy M., Haynes, Matthew, Kelley, Scott, Hong Liu, Mahaffy, Joseph M., Mueller, Jennifer E., Nulton, Jim, Olson, Robert, Parsons, Rachel, Rayhawk, Steve, Suttle, Curtis A., Rohwer, Forest. “The Marine Viromes of Four Oceanic Regions.” PLoS Biology 4 (2006): 2121-2131.
Azam, Farroq, and Alexandra Z. Worden. “Microbes, Molecules, and Ecosystems.” Science 303 (2004): 1622-1624.
Bertilsson, S., O. Berglund, D. M. Karl, and S. W. Chisholm. “Elemental composition of marine Prochlorococcus and Synechococcus: Implications for the ecological stoichiometry of the sea.” Limnology and Oceanography 48 (2003): 1721-731.
“Burkholderia.” Microbewiki: The Student-Edited Microbiology Resource. 16 August 2006. 20 August 2008 .
Carbon Cycling and Sequestration continued; Microbial Ocean Communities.” GTL. July-Aug. 2005. U.S. Department of Energy. 27 Aug. 2008.
CAMPBELL, L., H. A. NOLLA, AND D. VAULOT. 1994. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol. Oceanogr. 39: 954–961.
Carlowicz. “The Oceans Have Their Own Weather Systems.” Oceanus Magazine 45.1 (2006): 18-21.
Carlson, Craig J., Stephen J. Viocannoni, Dennis A. Hansell, Stuart J. Goldberg, Rachel Parsons, Mark P. Otero, Kevin Vergin, and Benjamin R. Wheeler. “Effect of nutrient amendments on bacterioplankton production, community structure, and DOC utilization in the northwestern Sargasso Sea.” Aquatic Microbial Ecology 30 (2002): 19-36.
Carpenter, Edward J., and K.L. Smith Jr. “Plastics on the Sargasso Sea Surface.” Science 175 (1972): 1240-241.
Casey, John R., Michael W. Lomas, Joanna Mandecki, and Donald E. Walker. “Prochlorococcus contributes to new production in the Sargasso Sea deep chlorophyll maximum.” Geophysical Research Letter 34 (2007).
“Changes in Ocean Conditions in Sargasso Sea Potential Cause for Decline in Eel Fishery.” NOAA. 6 Mar. 2008. United States Department of Commerce. 27 Aug. 2008 .
Chisholm, S. W., Frankel, S. L., Goericke, R., Olson, R. J., Palenik, B., Waterbury, J. B., West-Johnsrud, L. & Zettler, E. R. (1992) Arch. Microbiol. 157, 297–300.
Christie, P. J., K. Atmakuri, and V. Krishnamoorthy. “Biogenesis, architectural, and function of bacterial Type IV secretion systems.” Annual Review of Microbiology 59 (2005): 451-85.
Coston-Clements, L., L.R. Settle, D.E. Hoss and F.A. Cross. “Utilization of the Sargassum habitat by marine invertebrates and vertebrates – a review.” NOAA Technical Memorandum NMFS-SEFSC (1991): 296.
Cotner JB, Ammerman JW, Peele ER, Bentzen E (1997) Phosphorus- limited bacterioplankton growth in the Sargasso Sea. Aquat Microb Ecol 13:141–149
Cotter, Rosalind. “Marine Biology: Spawning Spot.” Nature 23 Feb. 2006: 926-26.
Dolowy, P., J. Mondzelewski, and R. Zawadzlea. “Cloning and characterization of a region responsible for the maintenance of megaplasmid pTAV3 of Paracoccus versutus UW1.” Plasmid 53 (2005): 239-50.
Dufresne, Alexis, Marcel Salanoubat, Frederic Partensky, Francois Artivuenave, Ilka M. Axmann, Valerle Barbe, Simone Duprat, Michael Y. Galperin, Eugene V. Koonin, Florence Le Gall, Kira S. Makarova, Martin Ostrowskl, Sophie Oztas, Catherine Robert, Igor B. Rogozin, David J. Scanlan, Nicole Tandeau de Marsac, Jean Welssenbach, Patick Wincker, Yuri L. Wolf, and Wolfgang R. Hess. “Genome sequence of the cyanobacterium Prochlorococcus marinus SS120,.” PNAS 100 (2003): 10020-0025
M. D. DuRand et al., Deep-Sea Res. Part II Top. Stud. Oceanogr. 48, 1983 (2001).
DURAND, M. D., R. J. OLSON, AND S. W. CHISHOLM. 2001. Phytoplankton population dynamics at the Bermuda Atlantic Timeseries station in the Sargasso Sea. Deep-Sea Res. II 48: 1983–2003.
Etnoyer, Peter. “Oasis in the Sargasso Sea.” Weblog entry. Deep Sea News Science Blogs. 23 May 2007. ScienceBlogs.com. 27 August 2008
Frazier, Marvin and J. Craig Venter. "Harnessing the power of genomics for energy and the environment." Industrial Biotechnology. March 1, 2005, 1(1): 6-8. doi:10.1089/ind.2005.1.6.
Francis, Christopher A., J. M. Beman, and Marcel M. Kuypers. “New processes and players in the nitrogen cycle: the microbial ecology of anaerobic and archaeal ammonia oxidation.” ISME 1 (2007): 19-27.
Genthe, Henry. “The Sargasso Sea”. Smithsonian 29 (1998): 82.
Ghedin, E., Claverie, J. M. “Mimivirus relatives in the Sargasso Sea.” Virology journal (2005): 62-69.
Giovannoni SJ, Rappé MS, Vergin K, Adair N (1996) 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria phylum. Proc Natl Acad Sci USA 93(7979–7984):7979–7984 Y
Giovanni, S. J., H. J. Tripp, and S. Givan. “Genome Streamlining in a Cosmopolitan Oceanic Bacterium.” Science 309 (2005): 1242-245.
Giovanni, Stephen. “Pelagibacter ubique: A perfect engineering solution?” 15 Apr. 2005. Monterey Bay Aquarium Research Institute. 24 Aug. 2008 .
Gladnick, P.J. “The Unique Sargasso Sea.” 2002. 28 Aug. 2008 .
Goericke, R. & Repeta, D. J. (1992) Limnol. Oceanogr. 37, 425–433.
Gonzalez, Jose M., Joseph S. Covert, and William B. Whitman. “Silicibacter pomeroyi sp. Nov. and Roseovarius nubinhibens sp. Nov., dimethylsulfoniopropriate-demethylating bacteria from marine environments.” International Journal of Systematic and Evolutionary Microbiology 53 (2003): 1261-269.
Greenpeace. “The trash vortex.” Greenpeace International. Date accessed: 14 Aug. 2008. http://www.greenpeace.org/international/campaigns/oceans/pollution/trash-vortex.
Sheean T. Haley and Sonya T. Dyhrman. “Phosphorus Scavenging in the Unicellular Marine Diazotroph Crocosphaera watsonii.” Applied and Environmental Microbiology 72 (2006): 1452-1458.
HERDMAN, M., R. W. CASTENHOLZ, J. B. WATERBURY, AND R. RIPPKA. 2001. Form-genus XIII. Synechococcus, p. 508–512. In D. R. Boone, R. W. Castenholz, and G. M. Garrity [eds.], Bergey’s manual of systematic bacteriology. V. 1, 2nd ed.
Hess, W. R., Rocap, G., Ting, C. S., Larimer, F., Stilwagen, S., Lamerdin, J. & Chisholm, S. W. (2001) Photosynth. Res. 70, 53–71.
Hooper, Sean D., Raes, Jeroen, Foerstner, Konrad U., Harrington, Eoghan D., Dalevi, Daniel, and Peer Bork. “A Molecular Study of Microbe Transfer between Distant Environments.” PLoS ONE. 2008 Jul 9; 3(7) :e2607 doi:10.1371/journal.pone.0002607
Kagan, Juliana, Sharon, Itai, Beja, Oded, and Jonathan C Kuhn. “The tryptophan pathway genes of the Sargasso Sea metagenome: new operon structures and the prevalence of non-operon organization.” Genome Biology (2008) 9: 1 27
Karner MB, Delong EF, Karl DM (2001) Archaeal dominance in the mesopelagic zone. Nature 409:507–510
Langlois, Rebecca J., Hümmer, Diana, and Julie LaRoche. “Abundances and Distributions of the Dominant nifH Phylotypes in the Northern Atlantic Ocean.” Applied and Environmental Microbiology 74 (2008): 1922-1931.
Lohmann, Kenneth J., Shaun D. Cain, Susan A. Dodge, and Catherine M. Lohmann. “Regional Magnetic Fields as Navigational Markers for Sea Turtles.” Science 294 (2001): 364-66.
Malmstrom Rex R., Kiene, Ronald P., Cottrell, Matthew T., and David L. Kirchman. “Contribution of SAR11 Bacteria to Dissolved Dimethylsulfoniopropionate and Amino Acid Uptake in the North Atlantic Ocean.” Applied and Environmental Microbiology 70 (2004): 4129-4135.
Malmstrom, Rex R., Ronald P. Kiene, Matthew T. Cottrell, and David L. Kirchman. “Contribution of SAR11 Bacteria to Dissolved Dimethylsulonioproprionate and Amino Acid Uptake in the North Atlantic.” Applied and Environmental Microbiology 70 (2004): 4129-135.
Man, D., Wang, W., Sabehi, G., Aravind, L., Post, A.F., Massana,R. “Diversification and spectral tuning in marine proteorhodopsins.” EMBO 22 (2003): 1725-1731.
McCord, John W. “American Eel.” South Carolina Department of Natural Resources. 27 Aug. 2008 .
Milton, Sarah, and Peter Lutz. Oil and Sea Turtles; Biology, Planning, and Response. NOAA. Aug. 2003.
Mistery of the Sargasso Sea Solved By LiveScience Staff. http://www.livescience.com/strangenews/070522_sargasso_sea.html.
Moore, L.R., R. E. GOERICKE, AND S. W. CHISHOLM. 1995. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 116: 259–275.
Moore, Lisa R., Anton F. Post, Gabrielle Rocap, and Sallie W. Chisholm. “Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus.” Limnology and Oceanography 47 (2002): 989-96.
Moran, Mary Ann, González, José M., Kiene, Ronald P. “Linking a Bacterial Taxon to Sulfur Cycling in the Sea: Studies of the Marine Roseobacter Group.” Geomicrobiology Journal 20 (2003): 375.
Moran, Mary A., Alison Buchan, and Jose M. Gonzalez. “Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment.” Nature 432 (2004): 910-13.
Moran, Mary A., and V. Armbrust. “Genomes of Sea Microbes.” Oceanography 20 (2007): 47-55.
Moran, Mary A. and E. Virginia Armbrust. “Genomes of Sea Microbes.” Oceanography 20 (2007): 47-55.
Murphy, Jamie. “The Perils of Plastic Pollution.” TIME 2 June 1986. 27 Aug. 2008. http://www.time.com/time/magazine/article/0,9171,961534,00.html.
Not. F., Gausling, R., Azam, F., Heldelberg, J.F., Worden, A.Z. “Vertical distribution if picoeukaryotic diversity in the Sargasso Sea.” Environmental Microbiology 9 (2007): 1233-1252.
R. J. Olson et al., Deep-Sea Res. Part I Oceanogr. Res. Pap. 37, 1033 (1990).
O’Malley, Maureen A. "The Nineteenth Century Roof of ‘everything is everywhere’." Nature Review Microbiology. 5 August 2007 647-651. http://www.nature.com/nrmicro/journal/v5/n8/abs/nrmicro1711.html
Orandorff, S.A., Colwell, R.R. “Distribution and identification of luminous bacteria from the Sargasso sea.” Applied and Environmental Microbiology (1980): 983-987.
PALINSKA, K. A., T. JAHNS, R. RIPPKA, AND N. TANDEAU DE MARSAC. 2000. Prochlorococcus marinus strain PCC 9511, a picoplanktonic cyanobacterium, synthesizes the smallest urease. Microbiology 146: 3099–3107.
PARTENSKY, F., W. R. HESS, AND D. VAULOT. 1999. “Prochlorococcus,a marine photosynthetic prokaryote of global significance.” Microbiol. Mol. Biol. Rev. 63: 106–127. C:N:P in Prochlorococcus and Synechococcus 1731
Patra, Jayanta Kumar, Rath, Sakti Kanta, Jena, Karmabeer, Rathod, Vijaya Kumar, Thatoi, Hrudayanath. “Evaluation of Antioxidant and Antimicrobial Activity of Seaweed (Sargassum sp.) Extract: A Study on Inhibition of Glutathione-S-Transferase Activity.” Turkish Journal of Biology 32 (2008): 119-125.
RIPPKA, R., AND OTHERS. 2000. Prochlorococcus marinus Chisholm et al. 1992, subsp. nov. pastoris, strain PCC9511, the first axenic chlorophyll a2/b2-containing cyanobacterium (Oxyphotobacteria). Int. J. Syst. Evol. Microbiol. 50: 1833–1847.
Rivkin RB, Anderson MR (1997) Inorganic nutrient limitation of oceanic bacterioplankton. Limnol Oceanogr 42:730–740
Ruder, Kate. "Scientists Discover One Million New Genes in Ocean Microbes." Genome News Network. 4 March 2004. 20 August 2008. http://www.genomenewsnetwork.org/articles/2004/03/04/sargasso.php.
Sabehi, G., Kirkup, B.C., Rozenberg, M., Stambler, N., Polz, M.F., Beja, O. “Adaptation and spectrum tuning in divergent marine proteorhodopsins from the eastern Mediterranean and the Sargasso Seas.” The ISME journal 1 (2007): 48-55.
Springer. KARL, D. M., AND F. C. DOBBS. 1998. Molecular approaches to microbial biomass estimation in the sea, p. 29–84. In K. Cooksey [ed.], Molecular approaches to the study of the ocean. Chapman & Hall.
Stingl, Ulrich, Tripp, Harry James, Giovannoni, Stephen J. “Improvements of high-throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site”. ISME Journal: Multidisciplinary Journal of Microbial Ecology 1 (2007): 361-371.
Sudek, Sebastian, Haygood, Margo G., Youssef, Diaa T. A., Schmidt, Eric W. “Structure of Trichamide, a Cyclic Peptide from the Bloom-Forming Cyanobacterium Trichodesmium erythraeum, Predicted from the Genome Sequence.” Applied and Environmental Microbiology 72 (2006): 4382-4387.
Sullivan, Matthew B., John B. Waterbury, and Sallie W. Chisholm. “Cyanophages infecting the oceanic caynobacterium Prochlorococcus.” Nature (2003): 1047-051. (Sullivan et al., 2003)
Tang, Xiaoting, Wei Yi, Gerhard R. Munske, Devi P. Adhikari, Natalia L. Zakharova, and James E. Bruce. “Profiling the Membrane Proteome of Shewanella oneidensis MR-1 with New Affinity Labeling Probes.” Journal of Proteome Research 6 (2006): 724-30.
Teal, John, and Mildred Teal. The Sargasso Sea. Boston, MA: Little Brown and Company, 1975. 3-5, 8-11, 16-17, 183-184.
“Prochlorococcus marinus NATL2A.” JGI. The Regents of the University of California. 27 Aug. 2008 .
“The Sargasso Sea.” 28 Aug. 2008 .
http://shs.westport.k12.ct.us/chia/caribbean/sargasso_sea.htm
“The Word SAR11.” New Scientist 177 (2003): 43.
Thingstad TF, Rassoulzadegan F (1995) Nutrient limitation, microbial food webs, and biological C-pumps: suggested interactions in a P-limited Mediterranean. Mar Ecol Prog Ser 117:299–306
Thingstad TF, Hagström Å, Rassoulzadegan F (1997) Accumulation of degradable DOC in surface waters: is it caused by a malfunctioning microbial loop? Limnol Oceanogr 42(2):398–404
Todd, J., R. Rodgers, and M. Wexler. “Structural and Regulatory Genes required to make the gas dimethyl sulfide in bacteria.” Science 315 (2007): 666-69.
Venter, J. Craig, Karin Remington, John F. Heidelberg, Aaron L. Halpern, Doug Rusch, Jonathan A. Eisen, Dongying Wu, Ian Paulsen, Karen E. Nelson, William Nelson, Derrick E. Fouts, Samuel Levy, Anthony H. Knap, Michael W. Lomas, Ken Nealson, Owen White, Jeremy Peterson, Jeff Hoffman, Rachel Parsons, Holly Baden-Tillson, Cynthia Pfannkoch, Yu-Hui Rogers, Hamilton O. Smith. “Environmental Genome Shotgun Sequencing of the Sargasso Sea.” Science 304 (2004): 66-74.
Watson, Paul. “The Plastic Sea.” The Tyee. 26 July 2006. 27 Aug. 2008. http://thetyee.ca/views/2006/07/26/plasticsea/.
Worden, Alexandra Z., and Brian J. Binder. “Application of dilution experiments for measuring.” Aquatic Microbial Ecology 30 (2003): 159-74.
Worden, A. Z., M. L. Cuvelier, and D. H. Bartlett. “In depth analyses of marine microbial community genomics.” Trends in Microbiology 14 (2006): 331-36.
Wu, Jingfeng, Sunda, William, Boyle, Edward A., Karl, David M. “Phosphate Depletion in the Western North Atlantic Ocean.” Science 289 (2000): 759.
WYMAN, M., R. P. F. GREGORY, AND N. G. CARR. 1985. Novel role for phycoerythrin in a marine cyanobacterium Synechococcus strain DC-2. Science 230: 818–820.
Yang, Chen, Dmitry A. Rodionov, Xiaoquing Li, Olga N. Laikova, Mikhail S. Gelfand, Olga P. Zagnitki, Margaret F. Romine, Anna Y. Obraztsova, Kenneth H. Nealson, and Andrei L. Osterman. “Comparative Genomics and Experimental Characterization of N-Acetylglucosamine Utilization Pathway of Shewanella oneidensis.” Journal of Biological Chemistry 281 (2006): 29872.
M. V. Zubkov et al., Prog. Oceanogr. 45, 369 (2000).
Edited by Sherry Pablo, Hugo Frazao, Patricia Tu, Asa Gardner, Cam Nguyen, HsiaoTing Wang, students of Rachel Larsen