Rice tungro bacilliform virus: Difference between revisions
| Line 18: | Line 18: | ||
[[Image:Tungro microscope.jpg]] | |||
Revision as of 22:34, 19 April 2009
Baltimore Classification
Viruses; Pararetro-transcribing viruses; Caulimoviridae; Tungrovirus;[NCBI link to find]
Species
Rice tungro bacilliform virus
|
NCBI: Taxonomy |
Description and Significance
Describe the appearance, habitat, etc. of the organism, and why you think it is important.
In 1975, RTBV was first recognized and then identified as a component member (along with Rice Tungro Spherical Virus) of the virus complex that is responsible for causing rice tungro disease. It is found to occur in Ortyza sativa and Ortyza sp. species of rice.
The virion consists of a capsid the is not enveloped. It is elongated and exhibits icosahedral symmetry. The capsid is bacilliform. The capsid shells of virions are composed of multiple layers. With a length of 110-400 nm and a width of 30-35 nm.
Rice tungro disease, the most important viral disease of rice, is widespread in South and Southeast Asia and is believed to be responsible for annual losses nearing 109 US dollars worldwide (Herdt 1991).
Rice tungro is a viral disease seriously affecting rice production in South and Southeast Asia. Tungro is caused by the simultaneous infection in rice of Rice tungro bacilliform virus (RTBV), a double-stranded DNA virus and Rice tungro spherical virus (RTSV), a single-stranded RNA virus.
RTBV and RTSV, also known as the ‘‘Tungro virus complex’’, are transmitted exclusively by the Green leafhopper, GLH, Nephotettix virescens
Tungrovirus Molecular biology VIRION
Non enveloped, bacilliform particles.
GENOME
Circular double stranded DNA of about 8000 base pairs with discontinuities in both strands: one in the transcribed strand and one to three in the non-transcribed strand.
Non enveloped, bacilliform particles.
GENOME
Circular double stranded DNA of about 8000 base pairs with discontinuities in both strands: one in the transcribed strand and one to three in the non-transcribed strand.
Genome Structure
Describe the size and content of the genome. How many chromosomes? Circular or linear? Other interesting features? What is known about its sequence?
Pararetroviruses have a DNA genome. The RTBV genome is a double stranded DNA of 8.0 kbp with two site-specific discontinuities resulting from replication by reverse transcriptase. It is circular and contains one molecule.
retroviruses have an RNA genome whereas pararetroviruses have a DNA genome; and, a proviral form of retroviruses is integrated into the host chromosome whereas the DNA of pararetroviruses
accumulates within the nucleus as multiple copies
of a circular chromosome 
RTBV genome is a double stranded DNA of 8.0 kbp with two site-specific discontinuities resulting from replication by reverse transcriptase [3,16]. The RTBV genome has four open reading frames (ORF)
Virion Structure, Metabolism and Life Cycle
Interesting features of cell structure; how it gains energy; what important molecules it produces.
The RTBV virion consists of an unenveloped capsid in a bacilliform particle shape. It is approximately 30nm in diameter and has a length of 130 nm. The virion contains circular DNA in a single molecule and a single coat protein species. 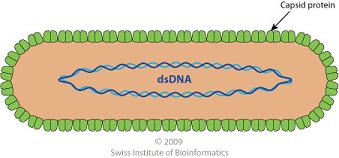
retroviruses have an RNA genome whereas pararetroviruses have a DNA genome; and, a proviral form of retroviruses is integrated into the host chromosome whereas the DNA of pararetroviruses accumulates within the nucleus as multiple copies of a circular chromosome
RTBV genome is a double stranded DNA of 8.0 kbp with two site-specific discontinuities resulting from replication by reverse transcriptase [3,16]. The RTBV genome has four open reading frames (ORF)
Rice tungro bacilliform virus (RTBV) replicates only in phloem cells in infected rice plants and its promoter drives strong phloem-specific reporter gene expression in transgenic rice plants. (http://images.google.com/imgres?imgurl=http://www.nature.com/emboj/journal/v16/n17/thumbs/7590500f5.jpg&imgrefurl=http://www.nature.com/emboj/journal/v16/n17/full/7590500a.html&usg=__8B6jGU34wMMqMKWlhViJo4xevbc=&h=193&w=150&sz=8&hl=en&start=14&tbnid=W6es5t8H8UfYAM:&tbnh=103&tbnw=80&prev=/images%3Fq%3DRice%2Btungro%2Bbacilliform%2Bvirus%26gbv%3D2%26hl%3Den%26safe%3Doff%26sa%)
Ecology and Pathogenesis
Habitat; symbiosis; biogeochemical significance; contributions to environment.
If relevant, how does this organism cause disease? Human, animal, plant hosts? Virulence factors, as well as patient symptoms.
References
Yanhai Yin2, Qun Zhu3, Shunhong Dai1, Chris Lamb3 and Roger N. Beachy1 "RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development "
Himani Tyagi Æ Shanmugam Rajasubramaniam Æ Manchikatla Venkat Rajam Æ.Indranil Dasgupta RNA-interference in rice against Rice tungro bacilliform virus results in its decreased accumulation in inoculated rice plants
Author
Page authored by _____, student of Prof. Jay Lennon at Michigan State University. A Viral Biorealm page on the family Rice tungro bacilliform virus
Baltimore Classification
Higher order taxa
Viruses; Pararetro-transcribing viruses; Caulimoviridae; Tungrovirus; Rice tungro bacilliform NCBI link to find]
Genera
Caulimovirus, Badnavirus, Soymovirus, Cavemovirus, Tungrovirus, Petuvirus
Description and Significance
All plant pararetroviruses are classified as members of the Caulimoviridae family. These viruses are similar to retroviruses, but instead of having an RNA genome, pararetroviruses have a DNA genome. (source: Marmey et al.)
Genome Structure
The genome structure of a caulimoviridae is either non-segmented or segmented. Because of this, the genome either consists of a single molecule or two segments of open circular double-stranded DNA. The length of the complete genome is 6800-7400-8175 nucleotides. There are terminally redundant sequences on the genome, which have direct terminal repeats. The genome has between one and eight open reading frames (ORFs). (sources: ICTVdB, Stavolone et al.)
Virion Structure of a Caulimoviridae
Caulimoviridae virions consist of a non-enveloped capsid. The capsid is isometric or bacilliform and exhibits icosahedral symmetry. The capsid has a diameter of 35-47.52-50nm and a length of 60-900nm. (source: ICTVdB)
Reproduction Cycle of a Caulimoviridae in a Host Cell
The replication of a caulimoviridae involves the reverse transcription of an RNA intermediate.
Viral Ecology & Pathology
Viruses in the Caulimoviridae family infect plants. Arthropods are the transmission vector. The viruses are distributed worldwide. (source: ICTVdB)
References
ICTVdB - The Universal Virus Database, version 3. http://www.ncbi.nlm.nih.gov/ICTVdb/ICTVdB/